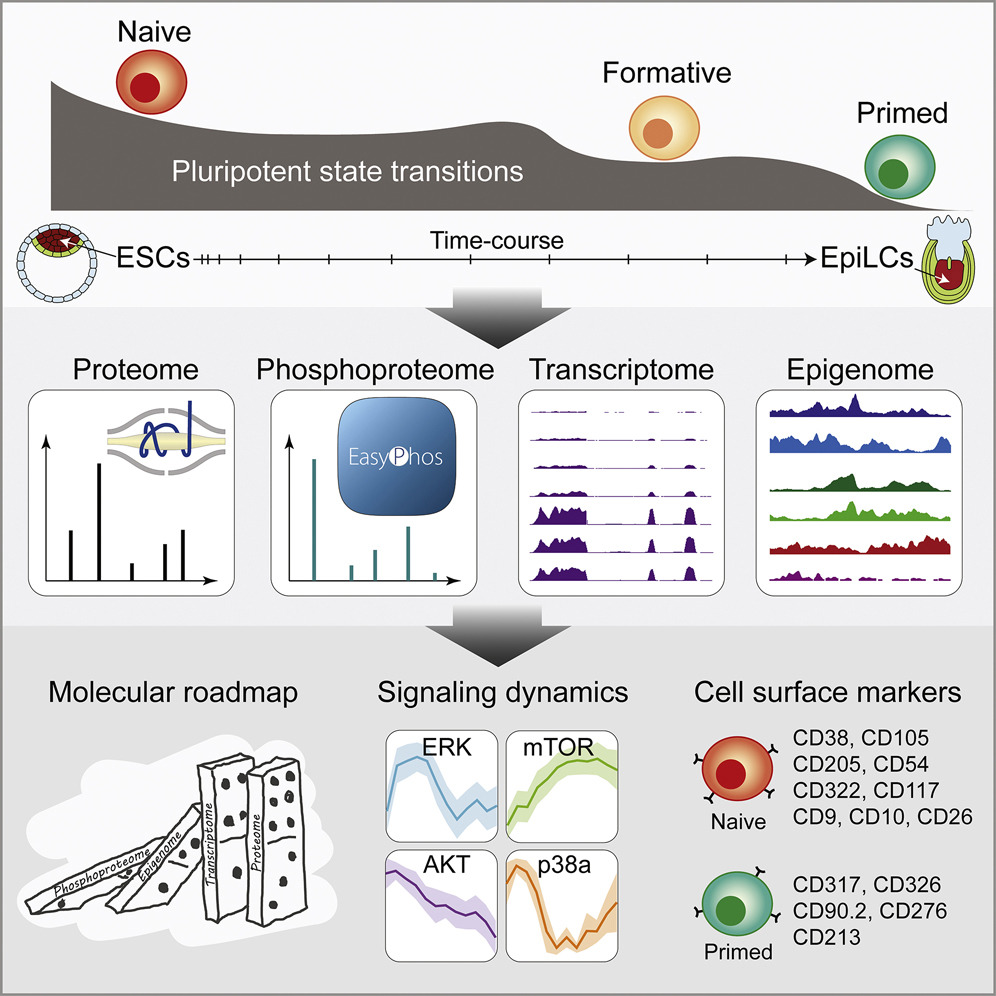
Pluripotency progression in stem cells and lineage specification
Pluripotency defines the potential of stem cells to differentiate into somatic and germ cells. The bookends of the pluripotency spectrum, naïve and primed states, are well studied but the progression of pluripotency from naïve to primed states remain to be fully characterised and their relationship with cell lineage specification remains to be investigated. We use multi-layered trans‐omics approach to study pluripotency transition from naïve to formative, an intermediate state, and then primed state. We developing methods that allows the reconstruction of the underlying trans-omic networks and correlating pluripotent transition to in vivo embryos during cell lineage specification.
Related publications:
Yang, P.✢†, Humphrey, S.✢†, Cinghu, S.✢, Pathania, R., Oldfield, A., Kumar, D., Perera, D., Yang, J., James, D., Mann, M. & Jothi, R.† (2019) Multi-omic profiling reveals dynamics of the phased progression of pluripotency. Cell Systems, 8(5), 427-445. [Full Text] [Biorxiv version] [The Stem Cell Atlas]
Kim, H., Osteil, P., Humphrey, S., Cinghu, S., Oldfield, A., Patrick, E., Wilkie, E., Peng, G., Suo, S., Jothi, R., Tam, P. & Yang, P.† (2020) Transcriptional network dynamics during the progression of pluripotency revealed by integrative statistical learning. Nucleic Acids Research, 48(4), 1828-1842. [Full Text]
Zheng, X., Yang, P., Lackford, B., Bennett, B., Wang, L., Li, H., Wang, Y., Miao, Y., Foley, J., Fargo, D., Jin, Y., Williams, C., Jothi, R. & Hu, G. (2016) CNOT3-dependent mRNA deadenylation safeguards the pluripotent state. Stem Cell Reports, 7(5), 897-910. [Text], [PDF]
Yang, P.✢†, Humphrey, S.✢†, Cinghu, S.✢, Pathania, R., Oldfield, A., Kumar, D., Perera, D., Yang, J., James, D., Mann, M. & Jothi, R.† (2019) Multi-omic profiling reveals dynamics of the phased progression of pluripotency. Cell Systems, 8(5), 427-445. [Full Text] [Biorxiv version] [The Stem Cell Atlas]
Kim, H., Osteil, P., Humphrey, S., Cinghu, S., Oldfield, A., Patrick, E., Wilkie, E., Peng, G., Suo, S., Jothi, R., Tam, P. & Yang, P.† (2020) Transcriptional network dynamics during the progression of pluripotency revealed by integrative statistical learning. Nucleic Acids Research, 48(4), 1828-1842. [Full Text]
Zheng, X., Yang, P., Lackford, B., Bennett, B., Wang, L., Li, H., Wang, Y., Miao, Y., Foley, J., Fargo, D., Jin, Y., Williams, C., Jothi, R. & Hu, G. (2016) CNOT3-dependent mRNA deadenylation safeguards the pluripotent state. Stem Cell Reports, 7(5), 897-910. [Text], [PDF]
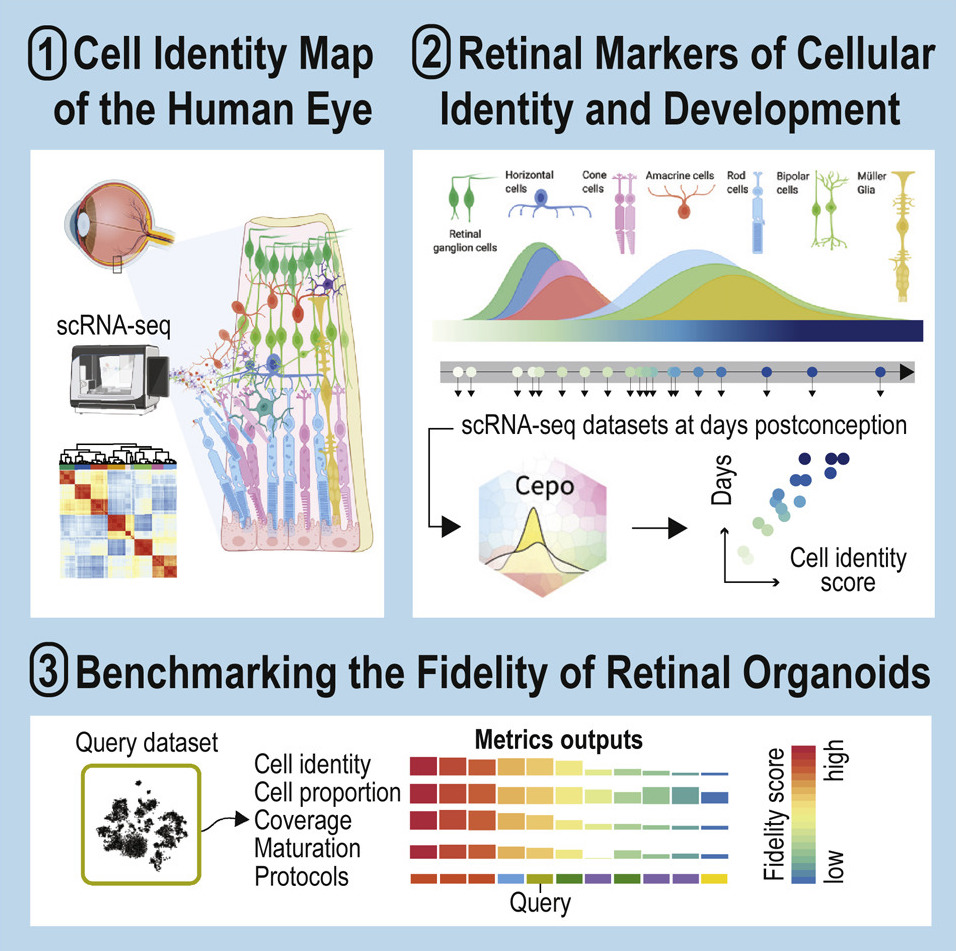
Stem cell and organoid engineering for regenerative medicine
A key goal of stem cell and organoid research is to generate stem cell-derived cells, tissues, and organs that can structurally and functionally replace any specific cell populations, tissues, and organs that are compromised by disease, injury, or ageing. While tremendous progress has been made in stem cell-based therapy, attempts to direct stem/progenitor cells to clinically useful cell types and tissues have often been fraught with imprecise and incomplete differentiation. We characterise stem cells and organoids using bulk and single-cell multi-omics approaches with the aim of realising their potential for regenerative medicine. Discovery made from these works will provide both basic and translational knowledge for stem cell and organoid engineering for tissue/organ regeneration.
Related publications:
Kim, H., O'Hara-Wright, M., Kim, D., Loi, T., Lim, B., Jamieson, R., Gonzalez-Cordero, A.† & Yang, P.† (2023) Comprehensive characterization of fetal and mature retinal cell identity to assess the fidelity of retinal organoids. Stem Cell Reports, 18(1), 175-189. [Full Text] [Eikon Shiny Web Server]
Fernando, M., Lee, S., Wark, J., Xiao, D., Lim, B., O'Hara-Wright, M., Kim, H., Smith, G., Wong, T., Teber, E., Ali, R., Yang, P., Graham, M. & Gonzalez-Cordero, A. (2022) Differentiation of brain and retinal organoids from confluent cultures of pluripotent stem cells connected by nerve-like axonal projections of optic origin. Stem Cell Reports, 17(6), 1476-1492. [Full Text]
Xiao, D., Caldow, M., Kim, H., Blazev, R., Koopman, R., Manandi, D., Parker, B. † & Yang, P.† (2022) Time-resolved phosphoproteome and proteome analysis reveals kinase signalling on master transcription factors during myogenesis. iScience, 25(6), 104489. [Full Text]
Kim, H., O'Hara-Wright, M., Kim, D., Loi, T., Lim, B., Jamieson, R., Gonzalez-Cordero, A.† & Yang, P.† (2023) Comprehensive characterization of fetal and mature retinal cell identity to assess the fidelity of retinal organoids. Stem Cell Reports, 18(1), 175-189. [Full Text] [Eikon Shiny Web Server]
Fernando, M., Lee, S., Wark, J., Xiao, D., Lim, B., O'Hara-Wright, M., Kim, H., Smith, G., Wong, T., Teber, E., Ali, R., Yang, P., Graham, M. & Gonzalez-Cordero, A. (2022) Differentiation of brain and retinal organoids from confluent cultures of pluripotent stem cells connected by nerve-like axonal projections of optic origin. Stem Cell Reports, 17(6), 1476-1492. [Full Text]
Xiao, D., Caldow, M., Kim, H., Blazev, R., Koopman, R., Manandi, D., Parker, B. † & Yang, P.† (2022) Time-resolved phosphoproteome and proteome analysis reveals kinase signalling on master transcription factors during myogenesis. iScience, 25(6), 104489. [Full Text]
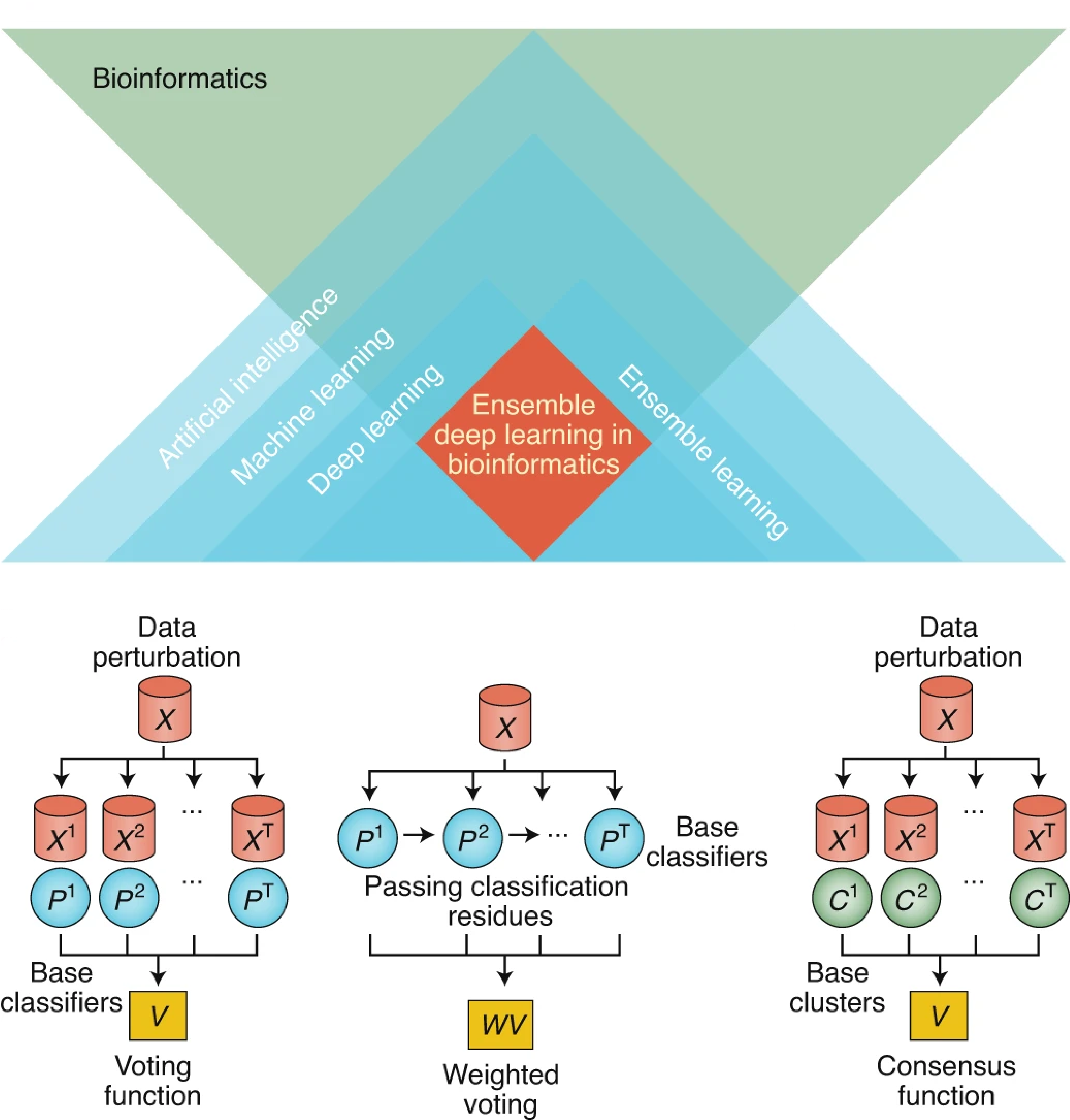
Machine learning for computational systems biology
Machine learning has become a key driver behind the revolution in computational systems biology. Leveraging on large-scale multi-omics data generated from high-througput biotechnological platforms, we develop specialised machine learning models, such as ensemble learning, deep learning, and multi-task learning models, to extract biological knowledge embedded in these data that are otherwise inaccessible. We aim to make machine learning more robust, flexiable, and interpretable for computational systems biology.
Related publications:
Cao, Y., Geddes, T., Yang, J. & Yang, P.† (2020) Ensemble deep learning in bioinformatics. Nature Machine Intelligence, 2, 500-508. [Full Text] [Nature Content Sharing link]
Geddes, T., Kim, T., Nan, L., Burchfield, J., Yang, J., Tao, D. & Yang, P.† (2019) Autoencoder-based cluster ensembles for single-cell RNA-seq data analysis. BMC Bioinformatics, 20, 660. [Full Text] [Repo]
Lin, Y., Cao, Y., Kim, H., Salim, A., Speed, T., Lin, D., Yang, P.† & Yang, J.† (2020) scClassify: sample size estimation and multiscale classification of cells using single and multiple reference. Molecular Systems Biology, 16, e9389. [Full Text] [BioC R package]
Cao, Y., Geddes, T., Yang, J. & Yang, P.† (2020) Ensemble deep learning in bioinformatics. Nature Machine Intelligence, 2, 500-508. [Full Text] [Nature Content Sharing link]
Geddes, T., Kim, T., Nan, L., Burchfield, J., Yang, J., Tao, D. & Yang, P.† (2019) Autoencoder-based cluster ensembles for single-cell RNA-seq data analysis. BMC Bioinformatics, 20, 660. [Full Text] [Repo]
Lin, Y., Cao, Y., Kim, H., Salim, A., Speed, T., Lin, D., Yang, P.† & Yang, J.† (2020) scClassify: sample size estimation and multiscale classification of cells using single and multiple reference. Molecular Systems Biology, 16, e9389. [Full Text] [BioC R package]
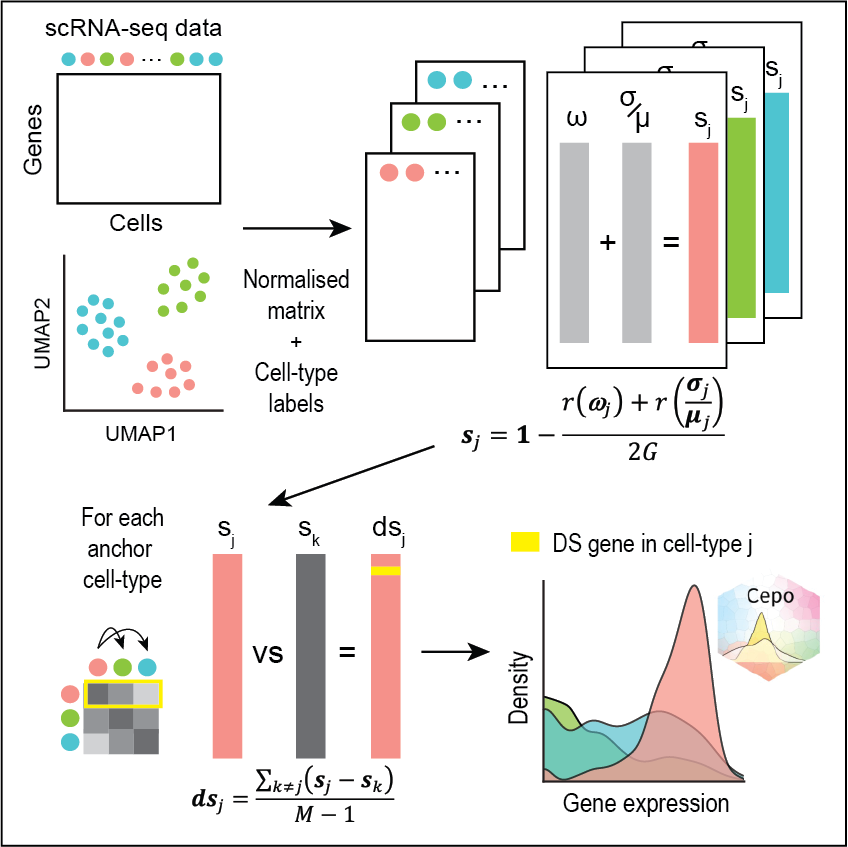
Data analytics for spatial and single-cell multi-omics
Data analytics for spatial and single-cell multi-omics refers to the analysis and integration of multiple types of genomic data (e.g. transcriptomics, epigenomics, and proteomics) generated from high-throughput single-cell or spatial technologies to gain a comprehensive understanding of cellular and spatiotemporal biological processes. The goal is to identify patterns and relationships between the different omics layers and to link them to cellular functions and phenotypes. We specialise in developing statistical methods and data analytics solutions such as dimensionality reduction, clustering, classification, and visualisation tools, as well as integration algorithms to combine the different omics data types into a unified view for characterising cellular and biological systems.
Related publications:
Kim, H., Wang, K., Chen, C., Lin, Y., Tam, PPL., Lin, D., Yang, J. & Yang, P.† (2021) Uncovering cell identity through differential stability with Cepo. Nature Computational Science, 1, 784-790. [Full Text] [Nature Content Sharing link] [BioC R package]
Kim, H.✢, Lin, Y.✢, Geddes, T., Yang, J. & Yang, P.† (2020) CiteFuse enables multi-modal analysis of CITE-seq data. Bioinformatics, 36(14), 4137-4143. [Full Text] [Biorxiv version] [BioC R package]
Kim, H.✢, Kim, T.✢, Hoffman, N., Xiao, D., James, D., Humphrey, S. & Yang, P.† (2021) PhosR enables processing and functional analysis of phosphoproteomic data. Cell Reports, 34(8), 108771. [Full Text] [BioC R package] [STAR Protocol]
Kim, H., Wang, K., Chen, C., Lin, Y., Tam, PPL., Lin, D., Yang, J. & Yang, P.† (2021) Uncovering cell identity through differential stability with Cepo. Nature Computational Science, 1, 784-790. [Full Text] [Nature Content Sharing link] [BioC R package]
Kim, H.✢, Lin, Y.✢, Geddes, T., Yang, J. & Yang, P.† (2020) CiteFuse enables multi-modal analysis of CITE-seq data. Bioinformatics, 36(14), 4137-4143. [Full Text] [Biorxiv version] [BioC R package]
Kim, H.✢, Kim, T.✢, Hoffman, N., Xiao, D., James, D., Humphrey, S. & Yang, P.† (2021) PhosR enables processing and functional analysis of phosphoproteomic data. Cell Reports, 34(8), 108771. [Full Text] [BioC R package] [STAR Protocol]
Transcriptional and epigenomic regulations in stem cells
Transcriptional regulation involves the activation or repression of gene expression through the binding of transcription factors and cofactors to specific and accessible genomics regions, while epigenomic regulation modulates such genomic accessibility through various changes such as the histone modifications (e.g. acetylation and methylation). In stem cells, transcriptional and epigenomic regulations are crucial for maintaining their pluripotency and differentiation potential. Disruptions in these regulatory mechanisms can therefore lead to stem cell dysfunction and the development of various diseases. We aim to understand the intricate interplay between transcriptional and epigenomic regulation in stem cells and during their differentiation into terminal cell types. Knowledge acquire from these research is crucial for the development of stem cell-based regenerative therapies.
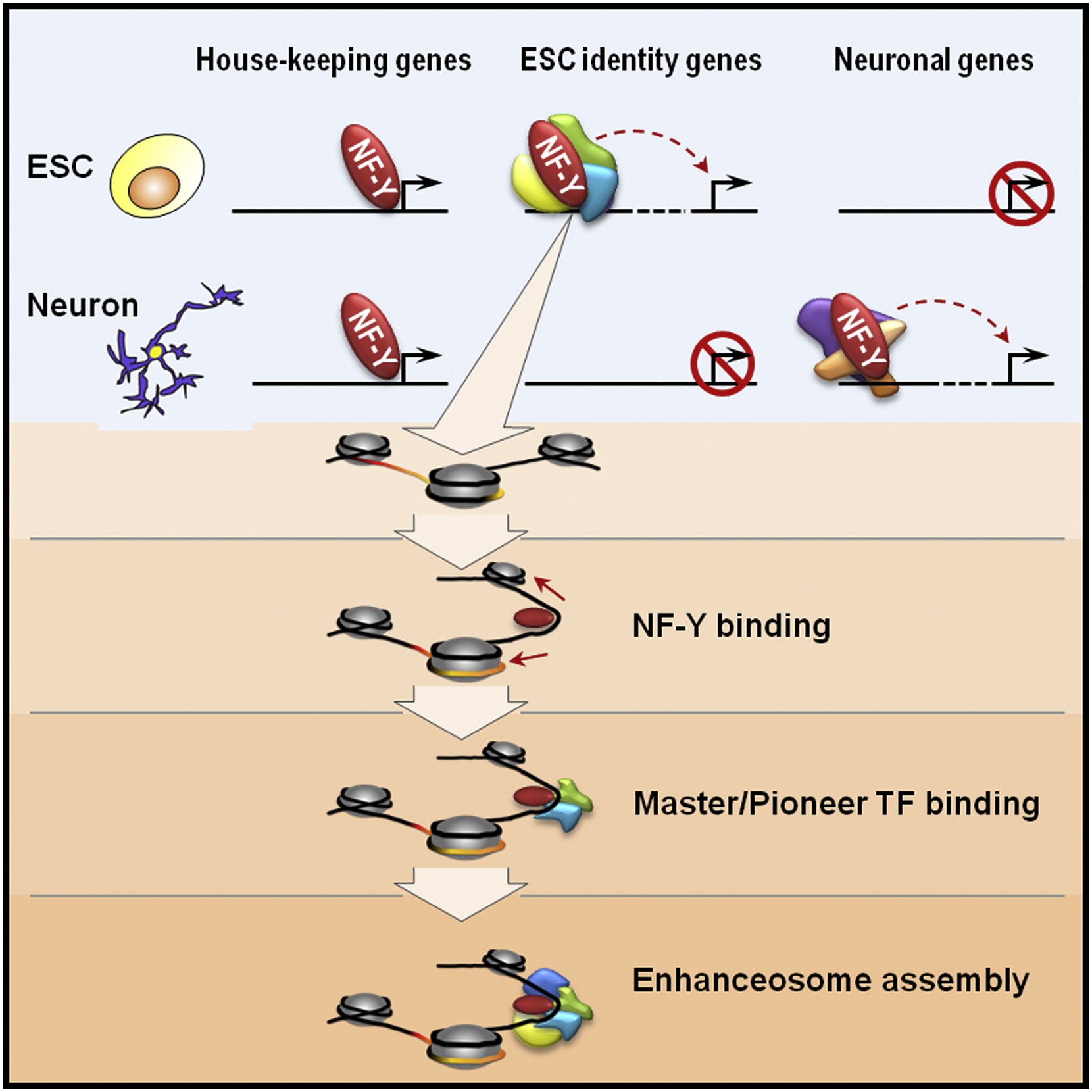
Related publications:
Oldfield, A.✢, Yang, P.✢, Conway, A., Cinghu, S., Freudenberg, J., Yellaboina, S. & Jothi, R. (2014). Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Molecular Cell, 55(5), 708-722. [Full Text], [PDF]
Kumar, D., Cinghu, S., Oldfield, A., Yang, P. & Jothi, R. (2021) Decoding the function of bivalent chromatin in development and cancer. Genome Research, 31(12), 2170-2184. [Full Text]
Cinghu, S.✢, Yang, P.✢, Kosak, J., Conway, A., Kumar, D., Oldfield, A., Adelman, K. & Jothi, R. (2017) Intragenic enhancers attenuate host gene expression. Molecular Cell, 68(1), 104–117. [Full Text], [PDF]
Oldfield, A.✢, Yang, P.✢, Conway, A., Cinghu, S., Freudenberg, J., Yellaboina, S. & Jothi, R. (2014). Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Molecular Cell, 55(5), 708-722. [Full Text], [PDF]
Kumar, D., Cinghu, S., Oldfield, A., Yang, P. & Jothi, R. (2021) Decoding the function of bivalent chromatin in development and cancer. Genome Research, 31(12), 2170-2184. [Full Text]
Cinghu, S.✢, Yang, P.✢, Kosak, J., Conway, A., Kumar, D., Oldfield, A., Adelman, K. & Jothi, R. (2017) Intragenic enhancers attenuate host gene expression. Molecular Cell, 68(1), 104–117. [Full Text], [PDF]