Notations:
Bold: CSB group member
✢: Co-first author
†: Corresponding/Co-corresponding author
Key publications

Multitask benchmarking of single-cell multimodal omics integration methods
Liu, C., Ding, S., Kim, H., Long, S., Xiao, D., Ghazanfar, S. & Yang, P.†
Nature Methods, https://doi.org/10.1038/s41592-025-02856-3. [Full Text], [Github of scripts and data], [Shiny Web Server]

Trans-omic profiling uncovers molecular controls of early human cerebral organoid formation
Chen, C., Lee, S., Zyner, K., Fernando, M., Nemeruck, V., Wong, E., Marshall, L., Wark, J., Aryamanesh, N., Tam, P., Graham, M.†, Gonzalez-Cordero, A.† & Yang, P.†
Cell Reports, 43(5), 114219 (2024). [Full Text], [Processed data at Github]
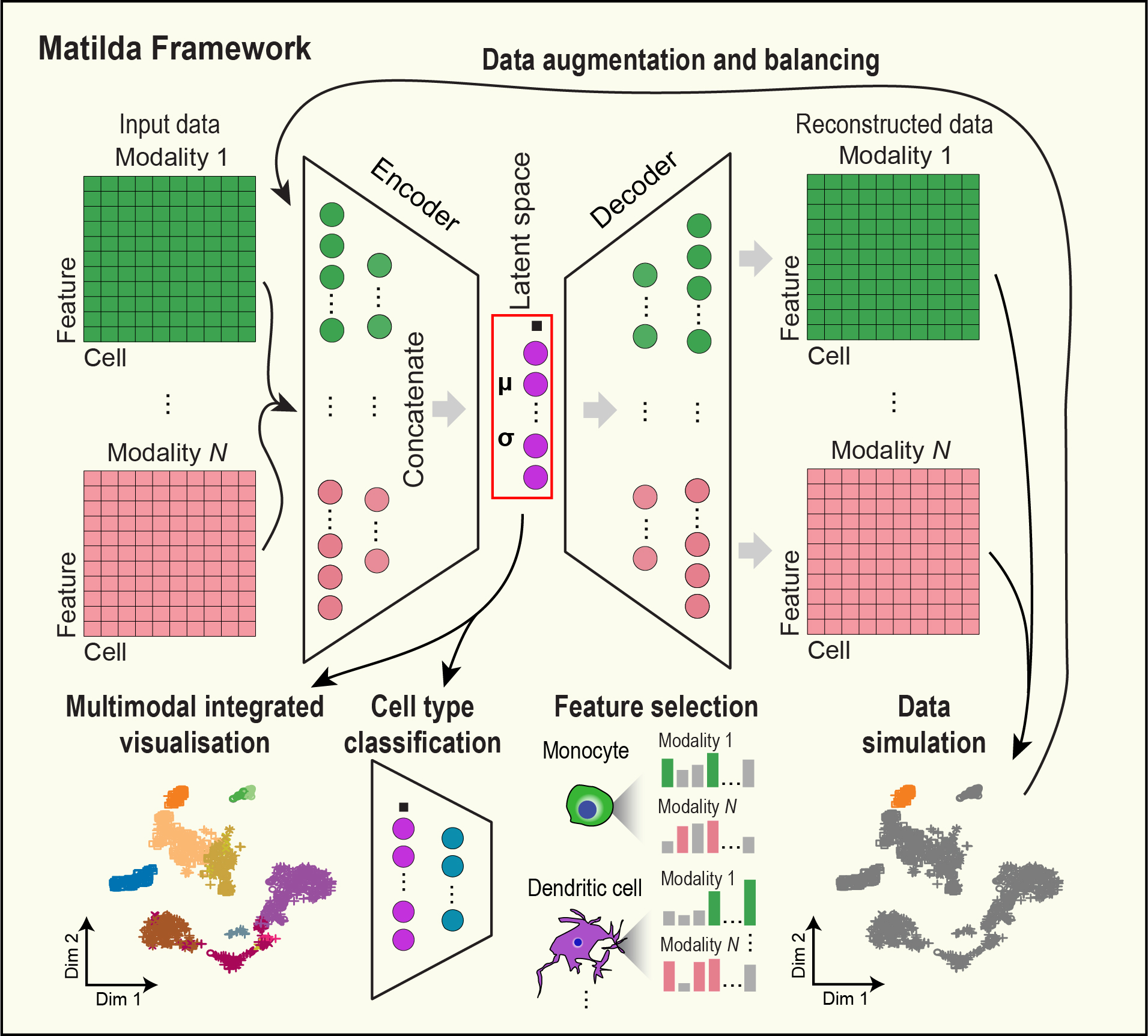
Multi-task learning from multimodal single-cell omics with Matilda
Liu, C., Huang, H. & Yang, P.†
Nucleic Acids Research, 51(8), e45 (2023). [Full Text], [Repo and tutorial]
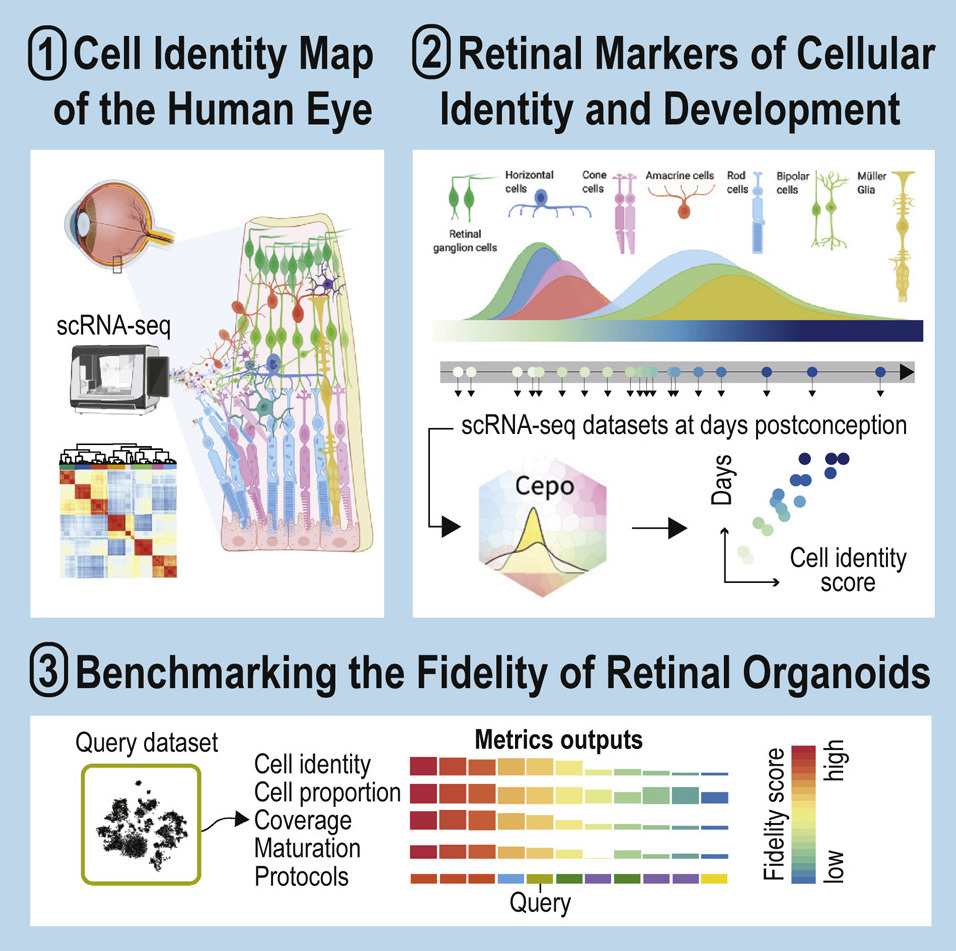
Comprehensive characterization of fetal and mature retinal cell identity to assess the fidelity of retinal organoids
Kim, H., O'Hara-Wright, M., Kim, D., Loi, T., Lim, B., Jamieson, R., Gonzalez-Cordero, A.† & Yang, P.†
Stem Cell Reports, 18(1), 175-189 (2023). [Full Text], [Eikon Shiny Web Server]
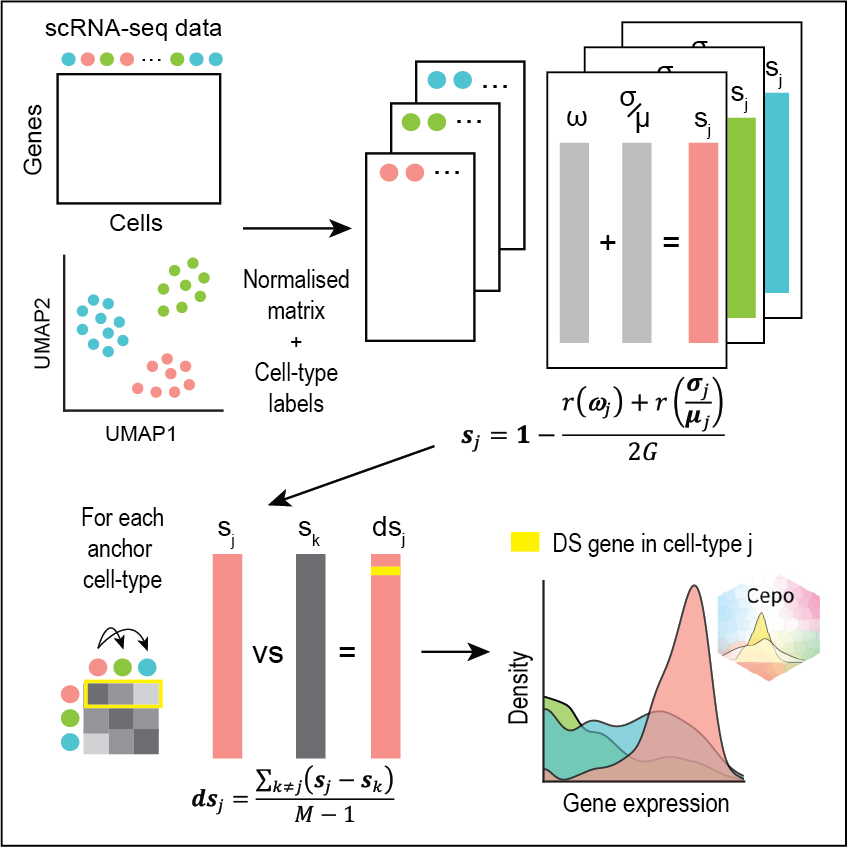
Uncovering cell identity through differential stability with Cepo
Kim, H., Wang, K., Chen, C., Lin, Y., Tam, PPL., Lin, D., Yang, J. & Yang, P.†
Nature Computational Science, 1, 784-790 (2021). [Full Text], [Nature Content Sharing link], [BioC R package]
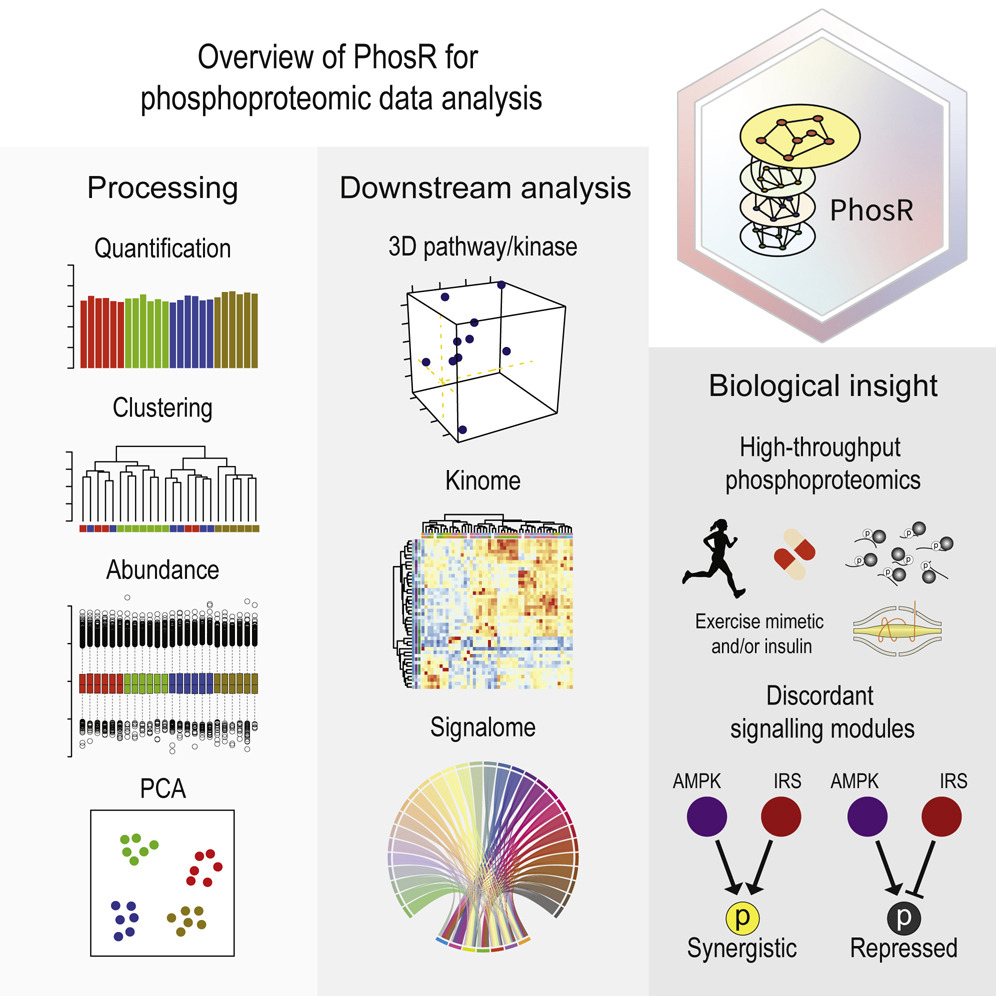
PhosR enables processing and functional analysis of phosphoproteomic data
Kim, H.✢, Kim, T.✢, Hoffman, N., Xiao, D., James, D., Humphrey, S. & Yang, P.†
Cell Reports, 34(8), 108771 (2021). [Full Text], [BioC R package]
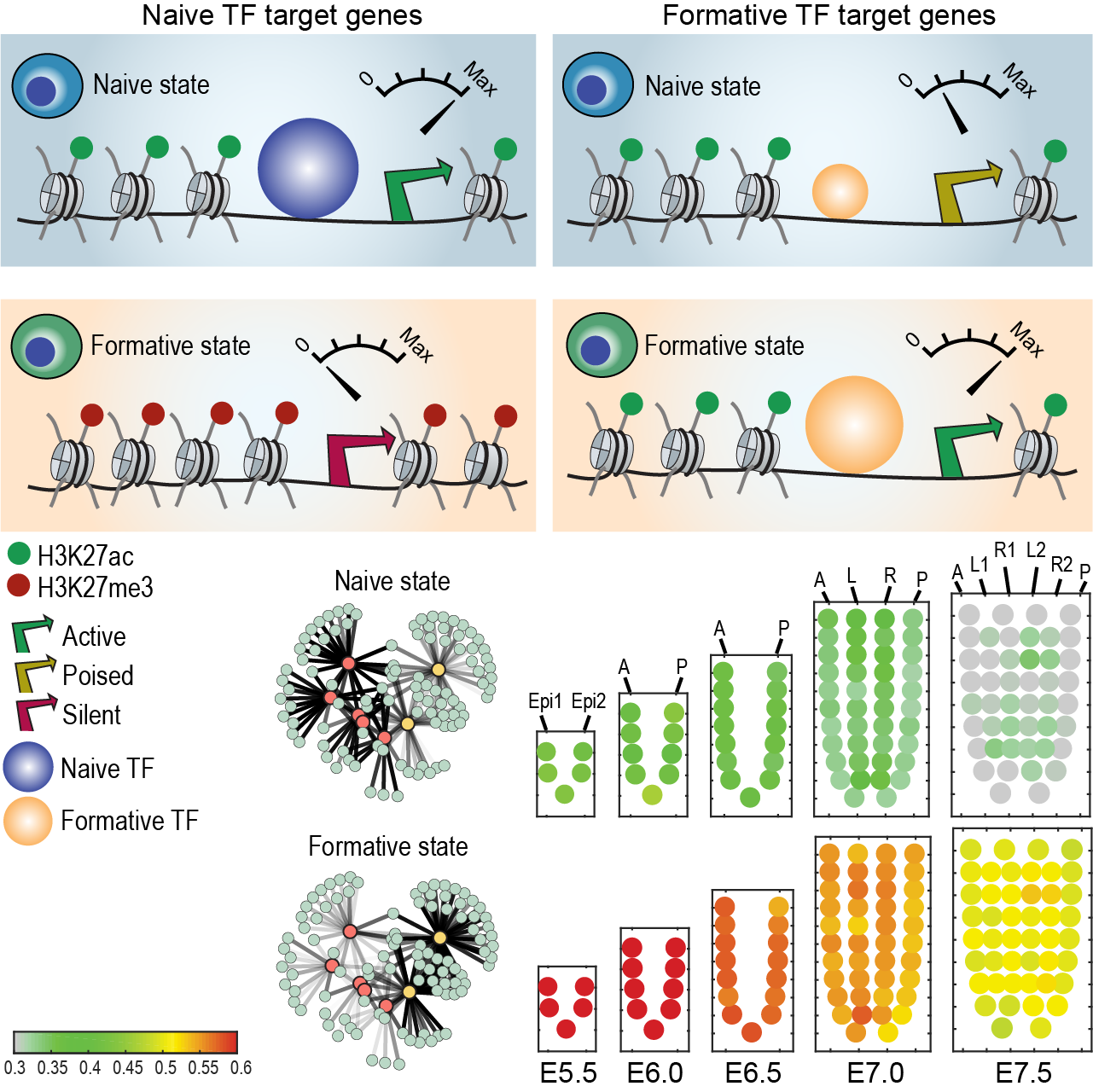
Transcriptional network dynamics during the progression of pluripotency revealed by integrative statistical learning
Kim, H., Osteil, P., Humphrey, S., Cinghu, S., Oldfield, A., Patrick, E., Wilkie, E., Peng, G., Suo, S., Jothi, R., Tam, P. & Yang, P.†
Nucleic Acids Research, 48(4), 1828-1842 (2020). [Full Text]
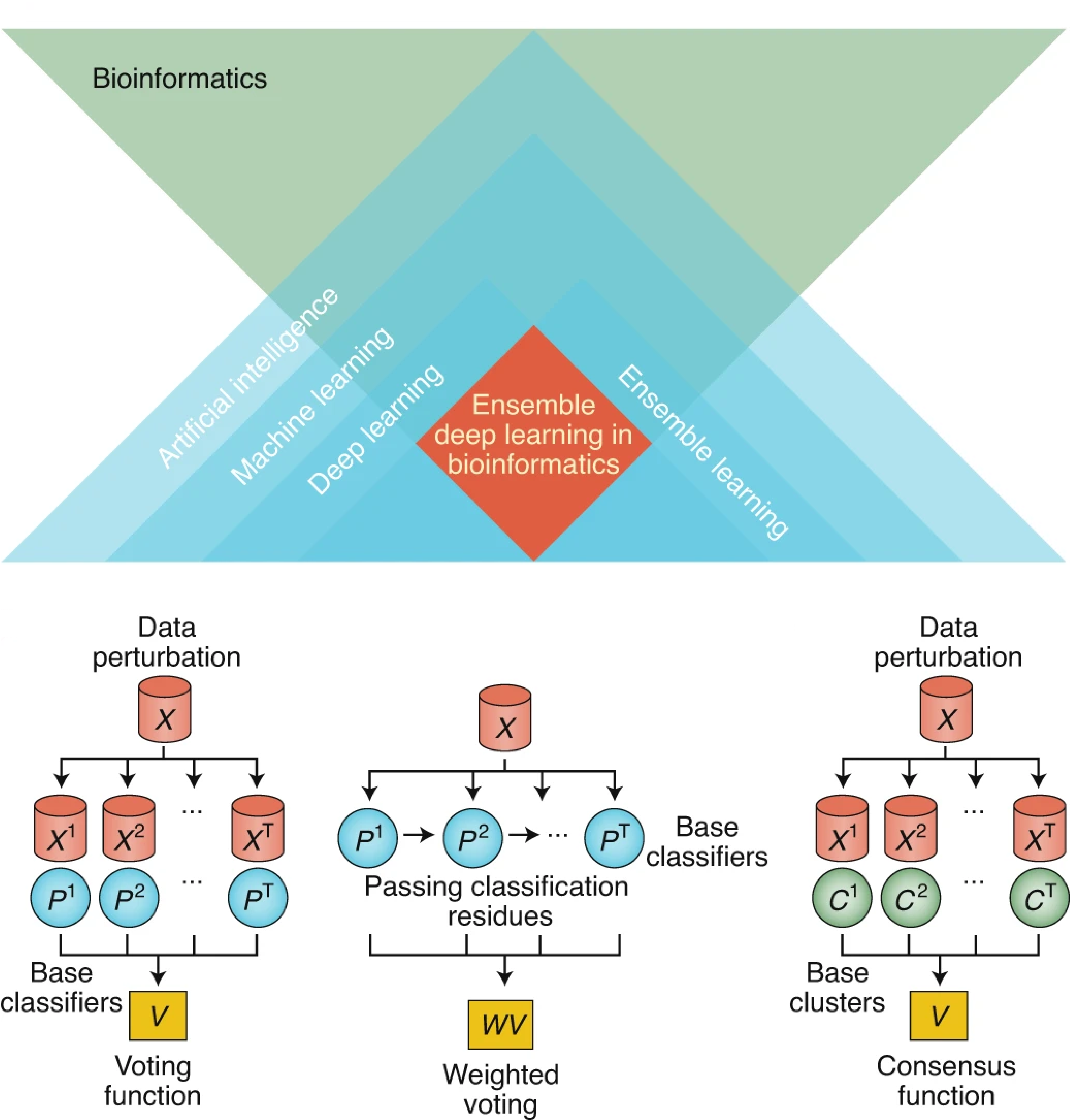
Ensemble deep learning in bioinformatics
Cao, Y., Geddes, T., Yang, J. & Yang, P.†
Nature Machine Intelligence, 2, 500-508 (2020). [Full Text], [Nature Content Sharing link]
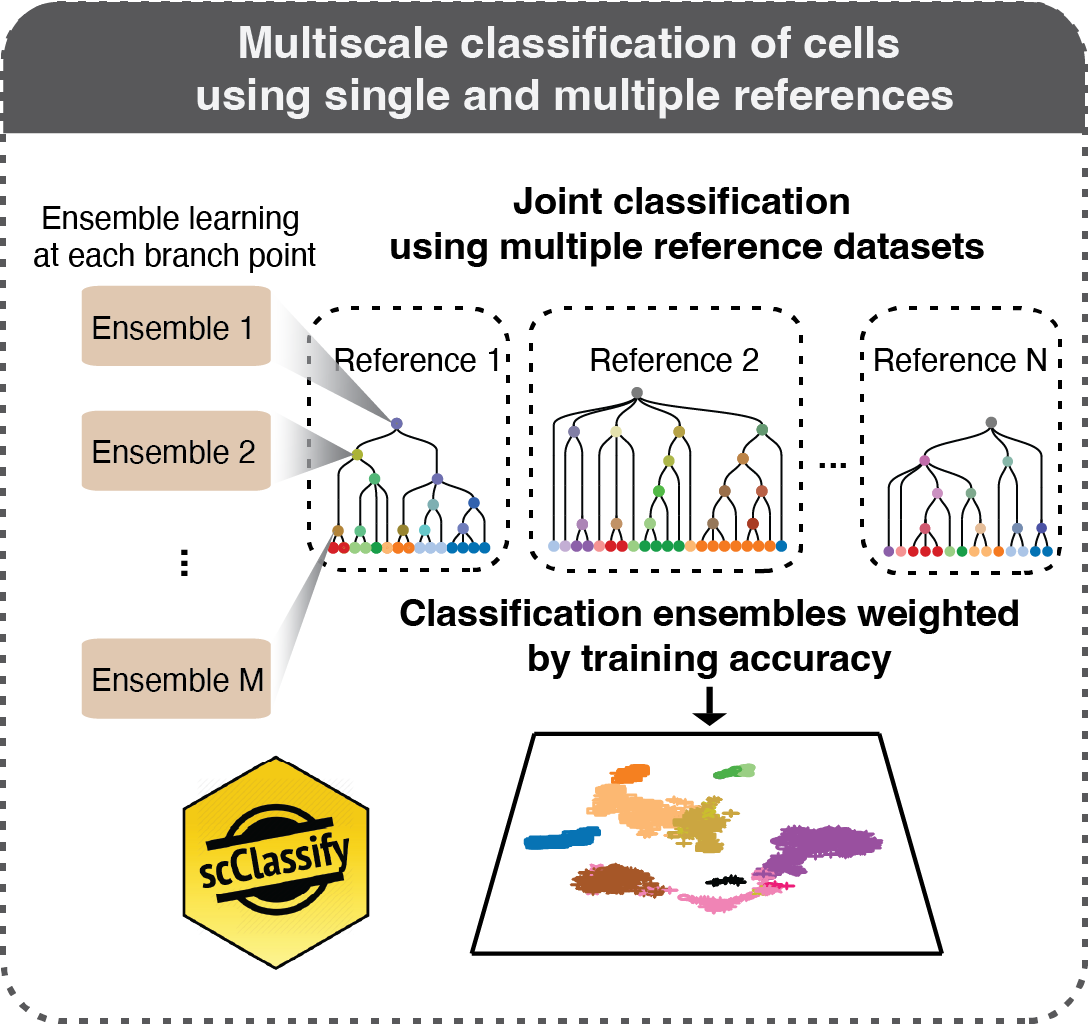
scClassify: sample size estimation and multiscale classification of cells using single and multiple reference
Lin, Y., Cao, Y., Kim, H., Salim, A., Speed, T., Lin, D., Yang, P.† & Yang, J.†
Molecular Systems Biology, 16, e9389 (2020). [Full Text], [BioC R package]
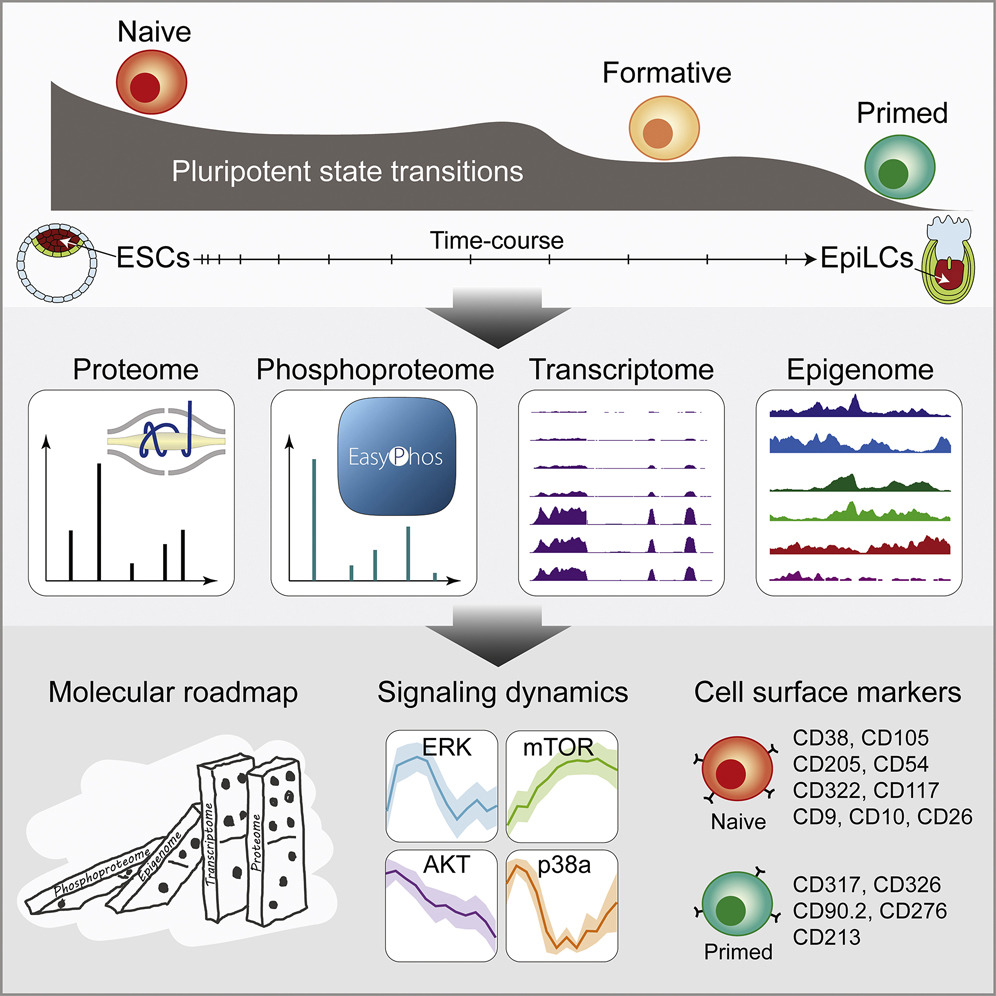
Multi-omic profiling reveals dynamics of the phased progression of pluripotency
Yang, P.✢†, Humphrey, S.✢†, Cinghu, S.✢, Pathania, R., Oldfield, A., Kumar, D., Perera, D., Yang, J., James, D., Mann, M. & Jothi, R.†
Cell Systems, 8(5), 427-445 (2019). [Full Text], [The Stem Cell Atlas]
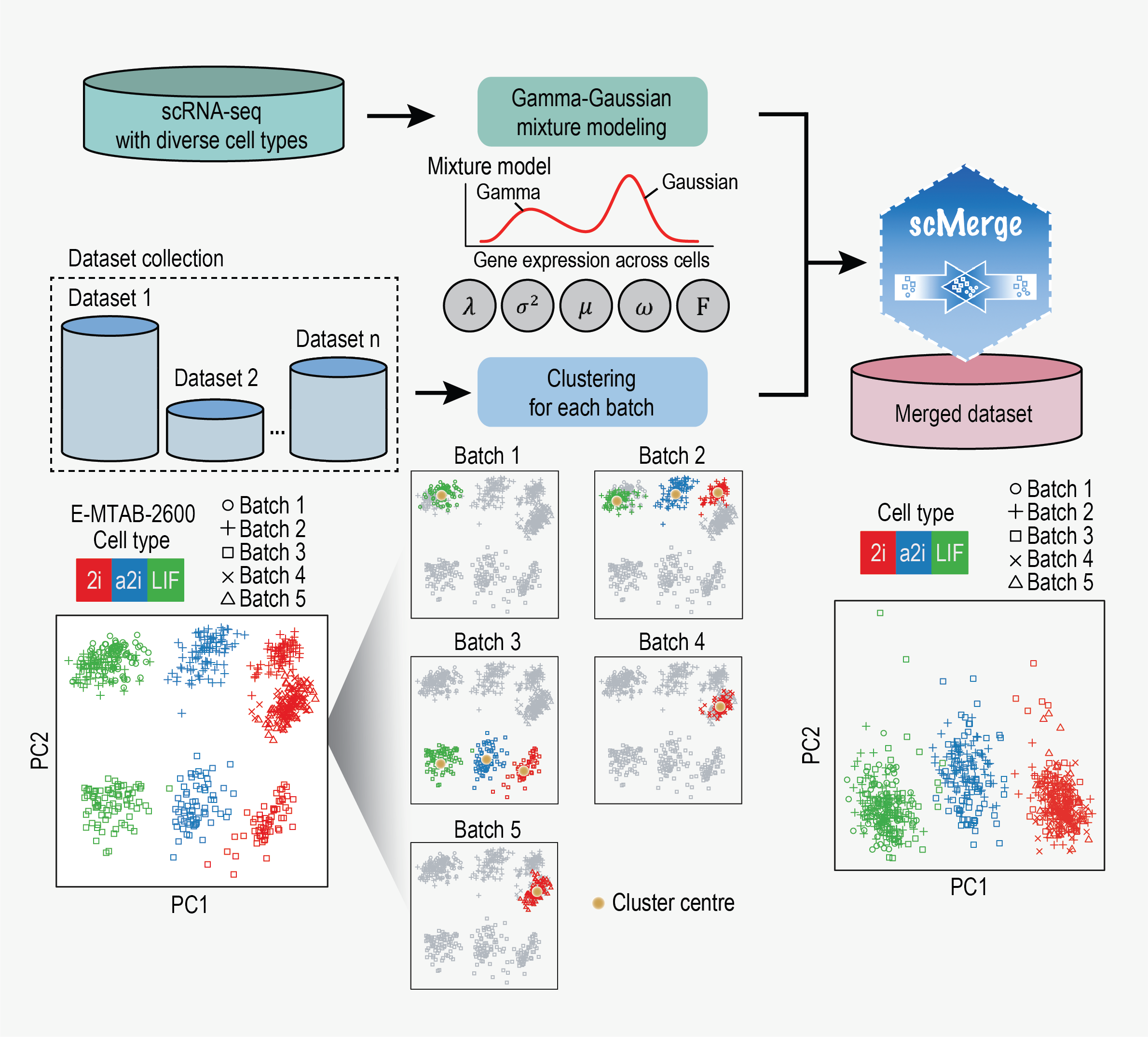
scMerge leverages factor analysis, stable expression, and pseudoreplication to merge multiple single-cell RNA-seq datasets
Lin, Y., Ghazanfar, S., Wang, K., Gagnon-Bartsch, J., Lo, K., Su, X., Han, Z., Ormerod, J., Speed, T., Yang, P.† & Yang, J.†
Proceedings of the National Academy of Sciences of the United States of America, 116(20), 9775-9784 (2019). [Full Text], [BioC R package]
All publications
2026
Systems biology:120. Dutt, M., Liao, L., Kim, H., Blazev, R., Chan, A., Kore, H., Bezawork-Geleta, A., Dong, L., Rivera, I., Wee, N., Molendijk, J., Wong, J., Haynes, V., Uribe, V., Lynch, G., Smith, K., Montgomery, M., Watt, M., Yang, P., Dodd, G., Vervoort, S., Sims, N. & Parker, B. (2026) Phosphoproteomics of aged insulin-resistant bone identifies P70S6K phosphorylation of AFF4 as a gene-specific transcriptional regulator. Nature Communications, 17, 1347. [Full Text]
2025
Systems biology:119. Joun, G., Kempe, E., Chen, B., Sterling, J., Abbassi, R., Friess, D., Singleton, M., Choudhury, C., Marian, O., du Preez, W., Recasens, A., Clark, T., Du, T., Low, J., Kim, H., Yang, P., Khor, J., Hoque, M., Indurthi, D., Kuchibhotla, M., Palanisamy, R., Jorgensen, W., Montgomery, A., Baker, J., Higginbottom, S., Tomaskovic-Crook, E., Crook, J., Loo, L., Day, B., Neely, G., Guccione, E., Johns, T., Kassiou, M., Feng, Y., Harris, L., Don, A. & Munoz, L. (2025) Histone methyltransferase PRDM9 promotes survival of drug-tolerant persister cells in glioblastoma. Nature Communications, 16, 10905. [Full Text]
118. Loi, T., Cheng, A., Kim, H., Fernando, M., Nash, B., Aryamanesh, N., Grigg, J., Yang, P., Gonzalez-Cordero, A. & Jamieson, R. (2025) Connecting cilium, stress response, and proteostasis abnormalities inform variant and therapy assessment in RPGRIP1 retinal organoids. Stem Cell Reports, 102717. [Full Text]
117. Toh, H., Xu, L., Chen, C., Yang, P., Sun, A.† & Ouyang, J.† (2025) BrainSTEM: A single-cell multiresolution fetal brain atlas reveals transcriptomic fidelity of human midbrain cultures. Science Advances, 11(44), eadu7944. [Full Text] [Online resource]
116. Masamsetti, V., Salehin, N., Kim, H., Santucci, N., Weatherstone, M., McMahon, R., Marshall, L., Knowles, H., Sun, J., Studdert, J., Aryamanesh, N., Wang, R., Jing, N., Yang, P., Osteil, P. & Tam, P. (2025) Lineage contribution of the mesendoderm progenitors in the gastrulating mouse embryo. Developmental Cell, 60(14), 1991-2006. [Full Text]
115. Hoffman, N., Whitfield, J., Xiao, D., Radford, B., Suni, V., Blazev, R., Yang, P., Parker, B. & Hawley, J. (2025) Phosphoproteomics uncovers exercise intensity-specific skeletal muscle signaling networks underlying high-intensity interval training in healthy male participants. Sports Medicine, 55(8), 1983-2004. [Full Text]
114. Shen, S., Werner, T., Lukowski, S., Andersen, S., Sun, Y., Shim, W., Mizikovsky, D., Kobayashi, S., Outhwaite, J., Chiu, H., Chen, X., Chapman, G., Martin, E., Xia, D., Pham, D., Su, Z., Kim, D., Yang, P., Tan, M., Sinniah, E., Zhao, Q., Negi, S., Redd, M., Powell, J., Dunwoodie, S., Tam, P., Boden, M., Ho, J., Nguyen, Q. & Palpant, N. (2025) Atlas of multilineage stem cell differentiationreveals TMEM88 as a developmentalregulator of blood pressure. Nature Communications, 16, 1356. [Full Text]
Methodology and tools:
113. Liu, C., Ding, S., Kim, H., Long, S., Xiao, D., Ghazanfar, S. & Yang, P.† (2025) Multitask benchmarking of single-cell multimodal omics integration methods. Nature Methods, 22, 2449-2460. [Full Text] [Repo] [Shiny server]
• highlighted in Nature Genetics, doi.org/10.1038/s41588-025-02437-2, 2025. [fulltext]
112. Kim, D., Ding, W., Shaw, A., Torkel, M., Turtle, C., Yang, P. & Yang, J. (2025) Multi-view gene panel characterization for spatially resolved omics. Briefings in Bioinformatics, 26(5), bbaf478. [Full Text]
111. Cao, Y., Yu, L., Torkel, M., Kim, S., Lin, Y., Yang, P., Speed, T., Ghazanfar, S. & Yang, J. (2025) The current landscape and emerging challenges of benchmarking single-cell methods. Briefings in Bioinformatics, 26(5), bbaf380. [Full Text]
110. Yu, L., Lin, X., Xu, X., Yang, P. & Yang, J. (2025) Interpretable Differential Abundance Signature (iDAS). Small Methods, e2500572. [Full Text]
109. Kim, D., Chen, C., Yu, L., Yang, J. & Yang, P.† (2025) CLUEY enables knowledge-guided clustering and cell type detection from sinlge-cell omics data. Bioinformatics, 41(10), btaf528. [Full Text] [Repo]
108. Salim, A., Bhuva, D., Chen, C., Tan, C., Yang, P., Davis, M. & Yang, J. (2025) SpaNorm: spatially-aware normalization for spatial transcriptomics data. Genome Biology, 26, 109. [Full Text]
2024
Systems biology:107. Chen, C., Lee, S., Zyner, K., Fernando, M., Nemeruck, V., Wong, E., Marshall, L., Wark, J., Aryamanesh, N., Tam, P., Graham, M.†, Gonzalez-Cordero, A.† & Yang, P.† (2024) Trans-omic profiling uncovers molecular controls of the early human cerebral organoid formation. Cell Reports, 43(5), 114219. [Full Text] [Repo]
106. Wu, Z., Shen, S., Mizikovsky, D., Cao, Y., Naval-Sanchez, M., Tan, S., Alvarez, Y., Sun, Y., Chen, X., Zhao, Q., Kim, D., Yang, P., Hill, T., Jones. A., Fairlie, D., Pébay, A., Hewitt, A., Tam, P., White, M., Nefzger, C. & Palpant, N. (2024) Wnt dose escalation during the exit from pluripotency identifies tranilast as a regulator of cardiac mesoderm. Developmental Cell, 59(6), 705-722. [Full Text]
Methodology and tools:
105. Wagle, M.✢, Long, S.✢, Chen, C., Liu, C. & Yang, P.† (2024) Interpretable deep learning in single-cell omics. Bioinformatics, 40(6), btae374. [Full Text]
104. Chen, C.✢, Kim, H.✢† & Yang, P.† (2024) Evaluating spatially variable gene detection methods for spatial transcriptomics data. Genome Biology, 25, 18. [Full Text] [Repo]
2023
Systems biology:103. Kim, H., O'Hara-Wright, M., Kim, D., Loi, T., Lim, B., Jamieson, R., Gonzalez-Cordero, A.† & Yang, P.† (2023) Comprehensive characterization of fetal and mature retinal cell identity to assess the fidelity of retinal organoids. Stem Cell Reports, 18(1), 175-189. [Full Text] [Eikon Shiny Web Server]
102. Kim, D., Tran, A., Kim, H., Lin, Y., Yang, J.† & Yang, P.† (2023) Gene regulatory network reconstruction: harnessing the power of single-cell multi-omic data. npj Systems Biology and Applications, 9, 51. [Full Text]
101. Ting, T., Coleman, P., Kim, H., Zhao, Y., Mulangala, J., Cheng, N., Li, W., Gunatilake, D., Johnstone, D., Loo, L., Neely, G., Yang, P., Götz, J., Vadas, M & Gamble, J. (2023) Vascular senescence and leak are features of the early breakdown of the blood–brain barrier in Alzheimer’s disease models. GeroScience, 45(6), 3307-3331. [Full Text]
100. Fazakerley, D., Gerwen, J., Cooke, K., Duan, X., Needham, E., Díaz-Vegas, A., Madsen, S., Norris, D., Shun-Shion, A., Krycer, J., Burchfield, J., Yang, P., Wade, M., Brozinick, J., James, D. & Humphrey, S. (2023) Phosphoproteomics reveals rewiring of the insulin signaling network and multi-nodal defects in insulin resistance. Nature Communications, 14, 923. [Full Text]
99. Xiao, D., Chen, C. & Yang, P.† (2023) Computational systems approach towards phosphoproteomics and their downstream regulation. Proteomics, 23(3-4), e2200068. [Full Text] [PDF]
Methodology and tools:
98. Huang, H., Liu, C., Wagle, M. & Yang, P.† (2023) Evaluation of deep learning-based feature selection for single-cell RNA sequencing data analysis. Genome Biology, 24, 259. [Full Text] [Repo]
97. Di, X., Lin, M., Liu, C., Geddes, T., Burchield, J, Parker, L., Humphrey, S. & Yang, P.† (2023) SnapKin: a snapshot deep learning ensemble for kinase-substrate prediction from phosphoproteomics data. NAR Genomics and Bioinformatics, 5(4), lqad099. [Full Text] [Repo]
96. Yu, L., Liu, C., Yang, J. & Yang, P.† (2023) Ensemble deep learning of embeddings for clustering multimodal single-cell omics data. Bioinformatics, 39(6), btad382. [Full Text] [Repo and tutorial]
95. Liu, C., Huang, H. & Yang, P.† (2023) Multi-task learning from multimodal single-cell omics with Matilda. Nucleic Acids Research, 51(8), e45. [Full Text] [Repo and tutorial]
94. Willie, E., Yang, P. & Patrick, E. (2023) The impact of similarity metrics on cell type clustering in highly multiplexed in situ imaging cytometry data. Bioinformatics Advances, 3(1), vbad141. [Full Text]
93. Patrick, E., Canete, N., Iyengar, S., Harman, A., Sutherland, G. & Yang, P. (2023) Spatial analysis for highly multiplexed imaging data to identify tissue microenvironments. Cytometry Part A, 103(7), 593-599. [Full Text]
92. Kim, T., Kim, H., Oldfield, A. & Yang, P. (2023) PAD2: interactive exploration of transcription factor genomic colocalization using ChIP-seq data. STAR Protocols, 4(2), 102203. [Full Text] [PAD2 Web Server]
91. Cao, Y., Ghazanfar, S., Yang, P. & Yang, J. (2023) Benchmarking of analytical combinations for COVID-19 outcome prediction using single-cell RNA sequencing data. Briefings in Bioinformatics, 24(3), bbad159. [Full Text]
90. Cao, Y., Tran, A., Kim, H., Robertson, N., Lin, Y., Torkel, M., Yang, P., Patrick, E., Ghazanfar, S. & Yang, J. (2023) Thinking process templates for constructing data stories with SCDNEY. F1000Research, 12, 261. [Full Text]
2022
Systems biology:89. Kim, H., Li, P., Kim, T., Oldfield, A., Zheng, X. † & Yang, P.† (2022) Integrative Analysis Reveals Histone Demethylase LSD1 Promotes RNA Polymerase II Pausing. iScience, 25(10), 105049. [Full Text]
88. Blazev, R., Carl, C., Ng, Y., Molendijk, J., Voldstedlund, C., Zhao, Y., Xiao, D., Kueh, A., Miotto, P., Haynes, V., Hardee, J., Chung, J., McNamara, J., Qian, H., Gregorevic, P., Oakhill, J., Herold, M., Jensen, T., Lisowski, L., Lynch, G., Dodd, G., Watt, M., Yang, P., Kiens, B., Richter, E., Parker, B. (2022) Phosphoproteomics of three exercise modalities identifies canonical signaling and C18ORF25 as an AMPK substrate regulating skeletal muscle function. Cell Metabolism, 34(10), 1561-1577. [Full Text]
87. Xiao, D., Caldow, M., Kim, H., Blazev, R., Koopman, R., Manandi, D., Parker, B. † & Yang, P.† (2022) Time-resolved phosphoproteome and proteome analysis reveals kinase signalling on master transcription factors during myogenesis. iScience, 25(6), 104489. [Full Text]
86. Yang, P. & Tam, P. (2022) Mouse organogenesis atlas at single-cell resolution. Cell, 185(10), 1625-1627. [Full Text]
85. Fernando, M., Lee, S., Wark, J., Xiao, D., Lim, B., O'Hara-Wright, M., Kim, H., Smith, G., Wong, T., Teber, E., Ali, R., Yang, P., Graham, M. & Gonzalez-Cordero, A. (2022) Differentiation of brain and retinal organoids from confluent cultures of pluripotent stem cells connected by nerve-like axonal projections of optic origin. Stem Cell Reports, 17(6), 1476-1492. [Full Text]
84. Xiao, D., Kim, H., Pang, I. & Yang, P.† (2022) Functional analysis of the stable phosphoproteome reveals cancer vulnerabilities. Bioinformatics, 38(7), 1956-1963. [Full Text] [List of stably phosphorylated sites]
83. Duan, X., Norris, D., Humphrey, S., Yang, P., Cooke, K., Bultitude, W., Parker, B., Conway, O., Burchfield, J., Krycer, J., Brodsky, F., James, D. & Fazakerley, D. (2022) Trafficking regulator of GLUT4-1 (TRARG1) is a GSK3 substrate. Biochemical Journal, 479(11), 1237-1256. [Full Text]
82. Tu, J., Li, W., Yang, S., Yang, P., Yan, Q., Wang, S., Lai, K., Bai, X., Wu, C., Ding, W., Cooper‐White, J., Diwan, A., Yang, C., Yang, H. & Zou, J. (2022) Single‐cell transcriptome profiling reveals multicellular ecosystem of nucleus pulposus during degeneration progression. Advanced Science, 9(3), 2103631. [Full Text]
Methodology and tools:
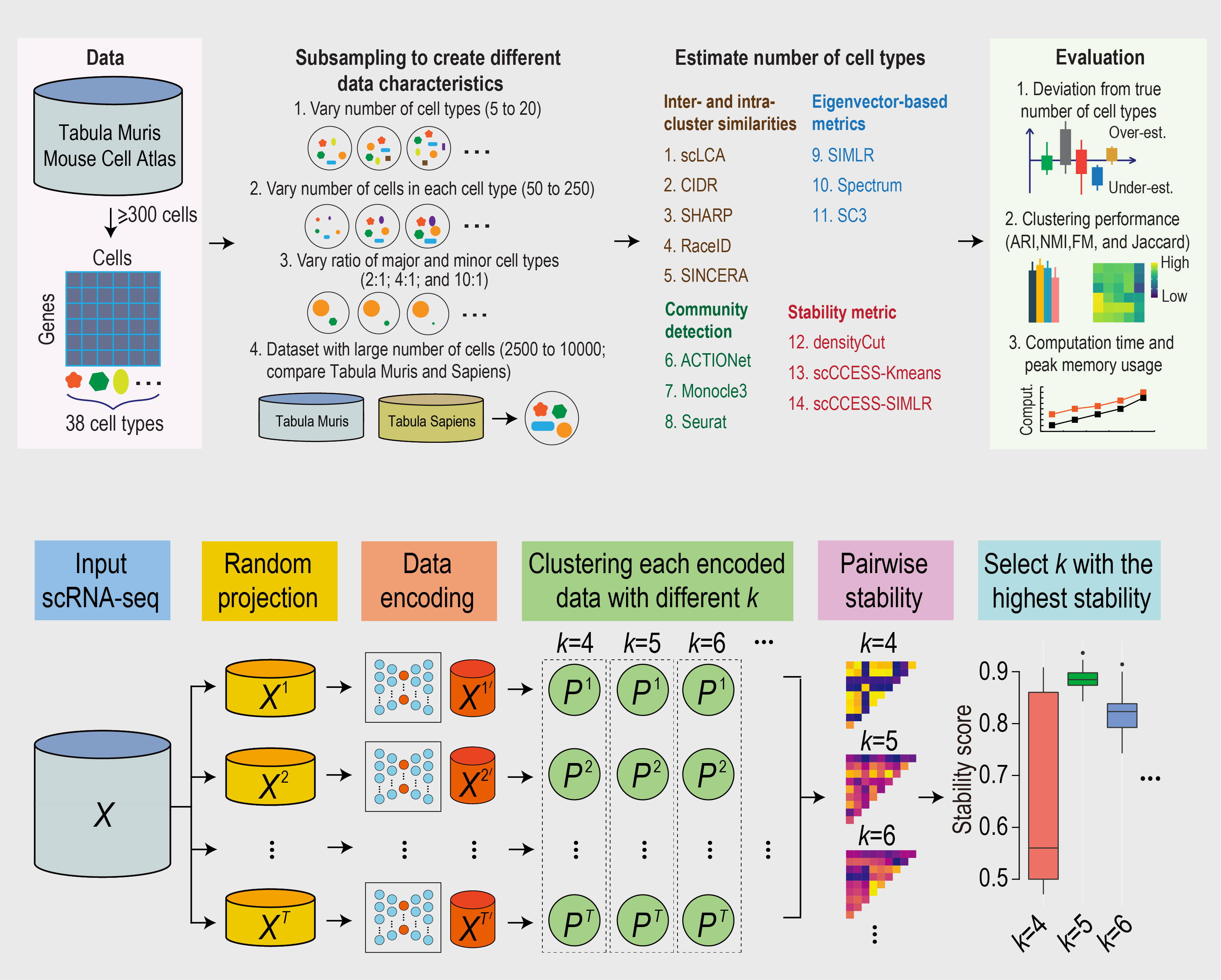
81. Yu, L., Cao, Y., Yang, J. & Yang, P.† (2022) Benchmarking clustering algorithms on estimating the number of cell types from single-cell RNA-sequencing data. Genome Biology, 23, 49. [Full Text] [Repo]
80. Cao, Y., Lin, Y., Patrick, E., Yang, P.† & Yang, J.† (2022) scFeatures: Multi-view representations of single-cell and spatial data for disease outcome prediction. Bioinformatics, 38(20), 4745-4753.
79. Tran, A., Yang, P., Yang, J. & Ormerod, J. (2022) Computational approaches for direct cell reprogramming: from the bulk omics era to the single cell era. Briefings in Functional Genomics, 21(4), 270-279. [Full Text]
78. Tran, A., Yang, P., Yang, J. & Ormerod, J. (2022) scREMOTE: Using multimodal single cell data to predict regulatory gene relationships and to build a computational cell reprogramming model. NAR Genomics and Bioinformatics, 4(1), lqac023. [Full Text]
2021
Systems biology:77. Kumar, D., Cinghu, S., Oldfield, A., Yang, P. & Jothi, R. (2021) Decoding the function of bivalent chromatin in development and cancer. Genome Research, 31(12), 2170-2184. [Full Text]
76. Kearney, A., Norris, D., Ghomlaghi, M., Wong, M., Humphrey, S., Carroll, L., Yang, G., Cooke, K., Yang, P., Geddes, T., Shin, S., Fazakerley, D., Nguyen, L., James, D. & Burchfield, J. (2021) Akt phosphorylates insulin receptor substrate to limit PI3K-mediated PIP3 synthesis. eLife, 10, e66942. [Full Text]
75. Kim, H., Tam, P. & Yang, P.† (2021) Defining cell identity beyond the premise of differential gene expression. Cell Regeneration, 10, 20. [Full Text]
74. Recasens, A., Humphrey, S., Ellis, M., Hoque, M., Abbassi, R., Chen, B., Longworth, M., Needham, E., James, D., Johns, T., Day, B., Kassiou, M., Yang, P. & Munoz, L. (2021) Global phosphoproteomics reveals DYRK1A regulates CDK1 activity in glioblastoma cells. Cell Death Discovery, 7, 81. [Full Text]
73. Norris, D.✢, Yang, P.✢, Shin, S.✢, Kearney, A., Kim, H., Geddes, T., Senior A., Fazakerley, D., Nguyen, L., James, D. & Burchfield, J. (2021) Signalling heterogeneity is defined by pathway architecture and inter-cellular variability in protein expression. iScience, 24(2), 102118. [Full Text]
72. Singh, R., Cottle, L., Loudovaris, T., Xiao, D., Yang, P., Thomas, H., Kebede M. & Thorn P. (2021) Enhanced structure and function of human pluripotent stemcell-derived beta-cells cultured on extracellular matrix. STEM CELLS Translational Medicine, 10, 492-505. [Full Text]
71. Su, X., Zhao, L., Shi, Y., Zhang, R., Long, Q., Bai, S., Luo, Q., Lin, Y., Zou, X., Ghazanfar, S., Tao, K., Yang, G., Wang, L., He, K., Cui, X., He, J., Wu, J., Han, B., Yan, B., Deng, B., Wang, N., Li, X., Yang, P., Hou, S., Sun, J., Yang, J., Chen, J. & Han, Z. (2021) Clonal evolution in liver cancer at single-cell and single-variant resolution. Journal of Hematology & Oncology, 14, 22. [Full Text]
70. Blazev, R., Ashwood, C., Abrahams, J., Chung, L., Francis, D., Yang, P., Watt, K., Qian, H., Quaife-Ryan, G., Hudson, J., Gregorevic, P., Thaysen-Andersen, M. & Parker, B. (2021) Integrated glycoproteomics identifies a role of N-glycosylation and galectin-1 on myogenesis and muscle development. Molecular & Cellular Proteomics, 20, 100030. [Full Text]
Methodology and tools:
69. Kim, H., Wang, K., Chen, C., Lin, Y., Tam, PPL., Lin, D., Yang, J. & Yang, P.† (2021) Uncovering cell identity through differential stability with Cepo. Nature Computational Science, 1, 784-790. [Full Text] [Nature Content Sharing link] [BioC R package]
68. Yang, P.†, Hao, H. & Liu, C. (2021) Feature selection revisited in the single-cell era. Genome Biology, 22, 321. [Full Text]
67. Cao, Y., Yang, P.† & Yang, J.† (2021) A benchmark study of simulation methods for single-cell RNA sequencing data. Nature Communications, 12, 6911. [Full Text] [Repo] [Shiny]
66. Kim, H., Kim, T., Xiao, D. & Yang, P.† (2021) Protocol for the processing and downstream analysis of phosphoproteomic data with PhosR. STAR Protocols, 2(2), 100585. [Full Text] [STAR Protocol]
65. Kim, H.✢, Kim, T.✢, Hoffman, N., Xiao, D., James, D., Humphrey, S. & Yang, P.† (2021) PhosR enables processing and functional analysis of phosphoproteomic data. Cell Reports, 34(8), 108771. [Full Text] [BioC R package] [STAR Protocol]
64. Kim, T., Tang, O., Vernon, S., Kott, K., Koay, Y., Park, J., James, D., Grieve, S., Speed, T., Yang, P.†, Figtree, G.†, O'Sullivan, J.† & Yang, J.† (2021) A hierarchical approach to removal of unwanted variation for large-scale metabolomics data. Nature Communications, 12, 4992. [Full Text] [Repo]
2020
Systems biology:63. Kim, H., Osteil, P., Humphrey, S., Cinghu, S., Oldfield, A., Patrick, E., Wilkie, E., Peng, G., Suo, S., Jothi, R., Tam, P. & Yang, P.† (2020) Transcriptional network dynamics during the progression of pluripotency revealed by integrative statistical learning. Nucleic Acids Research, 48(4), 1828-1842. [Full Text]
62. Azimi, A., Yang, P., Ali, M., Howard, V., Mann, G., Kaufman, K. & Fernandez-Penas, P. (2020) Data independent acquisition proteomic analysis can discriminate between actinic keratosis, Bowen’s disease and cutaneous squamous cell carcinoma. Journal of Investigative Dermatology, 140(1), 212–222. [Full Text]
Methodology and tools:
61. Cao, Y., Geddes, T., Yang, J. & Yang, P.† (2020) Ensemble deep learning in bioinformatics. Nature Machine Intelligence, 2, 500-508. [Full Text] [Nature Content Sharing link]
60. Lin, Y., Cao, Y., Kim, H., Salim, A., Speed, T., Lin, D., Yang, P.† & Yang, J.† (2020) scClassify: sample size estimation and multiscale classification of cells using single and multiple reference. Molecular Systems Biology, 16, e9389. [Full Text] [BioC R package]
59. Kim, H.✢, Lin, Y.✢, Geddes, T., Yang, J. & Yang, P.† (2020) CiteFuse enables multi-modal analysis of CITE-seq data. Bioinformatics, 36(14), 4137-4143. [Full Text] [Biorxiv version] [BioC R package]
58. Kaur, S., Peters, T., Yang, P., Luu, L., Vuong, J., Krycer, J. & O'Donoghue, S. (2020) Temporal ordering of omics and multiomic events inferred from time-series data. npj Systems Biology and Applications, 16, 22. [Full Text] [Repo]
57. Tang, T., Wu, H., Bao, W., Yang, P., Yuan, D., Zhou, B. (2020) New parallel algorithms for all pairwise computation on large HPC clusters. In the 20th International Conference on Parallel and Distributed Computing, Applications and Technologies (PDCAT), IEEE, 196-201. [Full Text]
• Best student paper award
2019
Systems biology:56. Su, Z.✢, Burchfield, J.✢, Yang, P.✢, Humphrey, S., Yang, G., Francis, D., Yasmin, S., Shin, S., Norris, D., Kearney, A., Astore, M., Scavuzzo, J., Fisher-Wellman, K., Wang, Q., Parker, B., Neely, G., Vafaee, F., Chiu, J., Yeo, R., Hogg, P., Fazakerley, D., Nguyen, L., Kuyucak, S. & James, D. (2019) Global redox proteome and phosphoproteome analysis reveals redox switch in Akt. Nature Communications, 10, 5486. [Full Text]
55. Yang, P.✢†, Humphrey, S.✢†, Cinghu, S.✢, Pathania, R., Oldfield, A., Kumar, D., Perera, D., Yang, J., James, D., Mann, M. & Jothi, R.† (2019) Multi-omic profiling reveals dynamics of the phased progression of pluripotency. Cell Systems, 8(5), 427-445. [Full Text] [Biorxiv version] [The Stem Cell Atlas]
54. Oldfield, A., Henriques, T., Kumar, D., Burkholder, A., Cinghu, S., Paulet, D., Bennett, B., Yang, P., Scruggs, B., Lavender, C., Rivals, E., Adelman, K. & Jothi, R. (2019) NF-Y controls fidelity of transcription initiation at gene promoters through maintenance of the nulceosome-depleted region. Nature Communications, 10, 3072. [Full Text]
53. Parker, B.✢, Calkin, A.✢, Seldin, M.✢, Keating, M. Tarling, E., Yang, P., Moody, S., Liu, Y., Zerenturk, E., Needham, E., Miller, M., Clifford, B., Morand, P., Watt, M., Meex, R., Peng, K., Lee, R., Jayawardana, K., Pan, C., Mellett, N., Weir, J., Lazarus, R., Lusis, A., Meikle, P., James, D., Vallim, T. & Drew, B. (2019) An integrative systems genetic analysis of mammalian lipid metabolism. Nature, 567, 187-193. [Full Text]
52. O'Sullivan, J., Neylon, A., Fahy, E., Yang, P., McGorrian, C. & Blake, G. (2019) MiR-93-5p is a novel predictor of coronary in-stent restenosis. Heart Asia, 11(1), e011134. [Full Text]
Methodology and tools:
51. Geddes, T., Kim, T., Nan, L., Burchfield, J., Yang, J., Tao, D. & Yang, P.† (2019) Autoencoder-based cluster ensembles for single-cell RNA-seq data analysis. BMC Bioinformatics, 20, 660. [Full Text] [Repo]
50. Cao, Y., Lin, Y., Ormerod, J., Yang, P., Yang, J. & Lo, K. (2019) scDC: single cell differential composition analysis. BMC Bioinformatics, 20, 721. [Full Text] Repo]
49. Kim, T., Lo, K., Geddes, T., Kim, H., Yang, J. & Yang, P.† (2019) scReClassify: post hoc cell type classification of single-cell RNA-seq data. BMC Genomics, 20, 913. [Full Text] [Repo]
48. Lin, Y., Ghazanfar, S., Strbenac, D., Wang, A., Patrick, E., Lin, D., Speed, T., Yang, J.† & Yang, P.† (2019) Evaluating stably expressed genes in single cells. GigaScience, 8(9), giz106. [Full Text] [Interactive tool for SEG download] [Giga DB] [human HK genes] [mouse HK genes]
47. Lin, Y., Ghazanfar, S., Wang, K., Gagnon-Bartsch, J., Lo, K., Su, X., Han, Z., Ormerod, J., Speed, T., Yang, P.† & Yang, J.† (2019) scMerge leverages factor analysis, stable expression, and pseudoreplication to merge multiple single-cell RNA-seq datasets. Proceedings of the National Academy of Sciences of the United States of America, 116(20), 9775-9784. [Full Text] [BioC R package]
46. Kim, T., Chen, I., Parker, B., Humphrey, S., Crossett, B., Cordwell, S., Yang, P.† & Yang, J.† (2019) QCMAP: An interactive web-tool for performance diagnosis and prediction of LC-MS Systems. Proteomics, 19(3), 1900068. [Full Text] [PDF] [QCMAP tool]
45. Yang, P.†, Ormerod, J., Liu, W., Ma, C., Zomaya, A. & Yang, J. (2019) AdaSampling for positive-unlabeled and label noise learning with bioinformatics applications. IEEE Transactions on Cybernetics, 49(5), 1932-1943. [PDF], [Repo]
44. Kim, T., Chen, I., Lin, Y., Wang, A., Yang, J. & Yang, P.† (2019) Impact of similarity metrics on single-cell RNA-seq data clustering. Briefings in Bioinformatics, 20(6), 2316-2326. [PDF] [draft] [Repo]
43. Ridder, M., Klein, K., Yang, J., Yang, P., Lagopoulos, J., Hickie, I., Bennett, M. & Kim, J. (2019) An uncertainty visual analytics framework for fMRI functional connectivity. Neuroinformatics, 17(2), 211-223. [PDF]
2018
Systems biology:42. Fazakerley, DJ., Chaudhuri, R., Yang, P., Maghzal, GJ., Thomas, KC., Krycer, JR., Humphrey, SJ., Parker, BL., Fisher-Wellman, KH., Meoli, CC., Hoffman, NJ., Diskin, C., Burchfield, JG., Cowley, MJ., Kaplan, W., Modrusan, Z., Kolumam, G., Yang, JYH, Chen, DL., Samocha-Bonet, D., Greenfield, JR., Hoehn, KL., Stocker, R. & James, DE. (2018) Mitochondrial CoQ deficiency is a common driver of mitochondrial oxidants and insulin resistance. eLife, 7, e32111. [Full Text]
2017
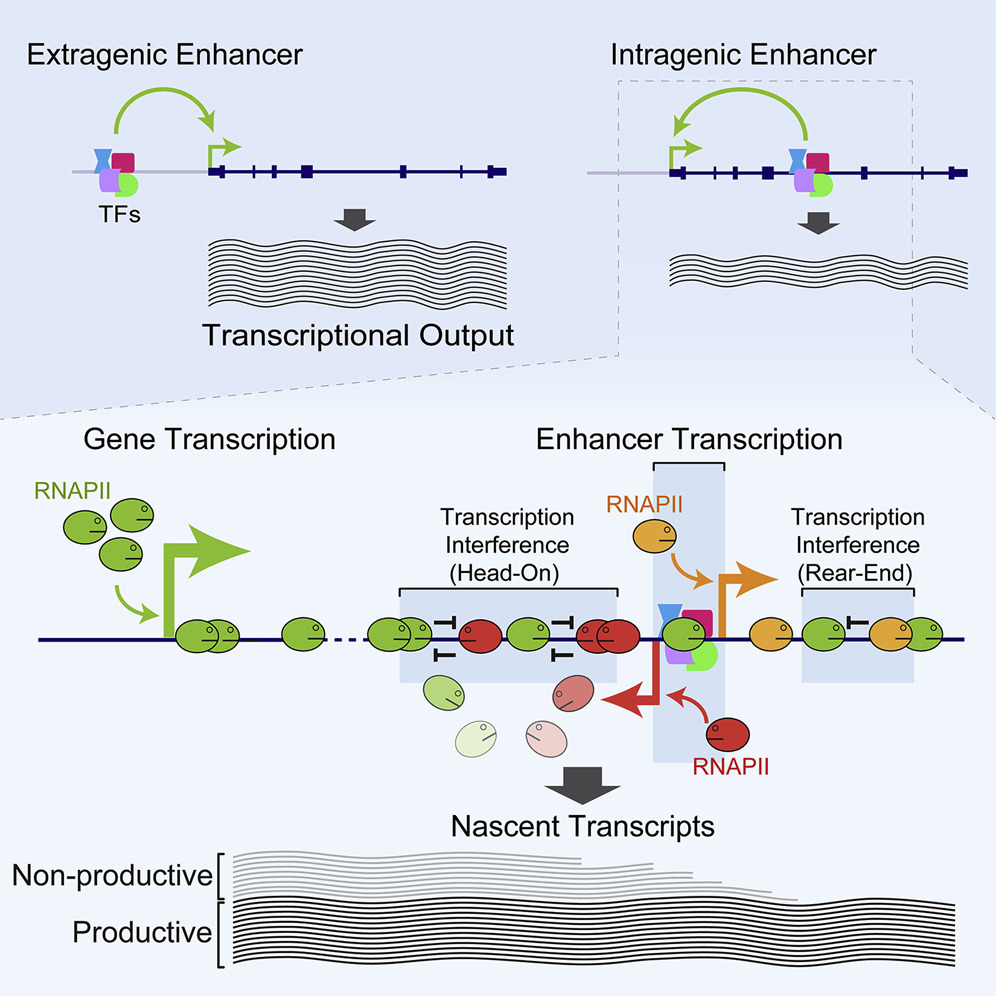
Systems biology:41. Cinghu, S.✢, Yang, P.✢, Kosak, J., Conway, A., Kumar, D., Oldfield, A., Adelman, K. & Jothi, R. (2017) Intragenic enhancers attenuate host gene expression. Molecular Cell, 68(1), 104–117. [Full Text], [PDF]
• highlighted in Nature Reviews Genetics, doi:10.1038/nrg.2017.90, 2017. [fulltext]
• also highlighted in Nature Reviews Molecular Cell Biology, doi:10.1038/nrm.2017.111, 2017. [fulltext]
40. Norris, DM.✢, Yang, P.✢, Krycer JR., Fazakerley DJ, James, DE. & Burchfield JG.† (2017) An improved Akt reporter reveals intra- and inter- cellular heterogeneity and oscillations in signal transduction. Journal of Cell Science, 130, 2757-2766. [PDF]
39. Yang, P.†, Oldfield, A., Kim, T., Yang, A., Yang, J. & Ho, J.† (2017) Integrative analysis identifies co-dependent gene expression regulation of BRG1 and CHD7 at distal regulatory sites in embryonic stem cells. Bioinformatics, 33(13), 1916–1920. [Full Text], [Interactive PAD]
Methodology and tools:
38. Yang, P., Liu, W. & Yang, J. (2017) Positive unlabeled learning via wrapper-based adaptive sampling. In Proceedings of the 26th International Joint Conference on Artificial Intelligence (IJCAI), Pre-print [PDF]
2016
Systems biology:37. Zheng, X., Yang, P., Lackford, B., Bennett, B., Wang, L., Li, H., Wang, Y., Miao, Y., Foley, J., Fargo, D., Jin, Y., Williams, C., Jothi, R. & Hu, G. (2016) CNOT3-dependent mRNA deadenylation safeguards the pluripotent state. Stem Cell Reports, 7(5), 897-910. [Text], [PDF]
36. Minard, A., Tan, S., Yang, P., Fazakerley, D., Domanova, W., Parker, B., Humphrey, S., Jothi, R., Stöckli. J. & James. D. (2016) mTORC1 is a major regulatory node in the FGF21 signaling network in adipocytes. Cell Reports, 17(1), 29-36. [Pubmed]
Methodology and tools:
35. Yang, P., Patrick, E., Humphrey, S., Ghazanfar, S., James, D., Jothi, R. & Yang, J. (2016). KinasePA: Phosphoproteomics data annotation using hypothesis driven kinase perturbation analysis. Proteomics, 16(13), 1868-1871. [Full Text], [Github Repo & tutorial], [CRAN R package]
34. Yang, P.†, Humphrey, S., James, D., Yang, J. & Jothi, R.† (2016). Positive-unlabeled ensemble learning for kinase substrate prediction from dynamic phosphoproteomics data. Bioinformatics, 32(2), 252-259. [Pubmed], [PDF], [Github Repo]
33. Lu, C., Wang, J., Zhang, Z., Yang, P. & Yu, G. (2016). NoisyGOA: Noisy GO annotations prediction using taxonomic and semantic similarity. Computational Biology and Chemistry, 65, 203-211.
32. Domanova, W., Krycer, J., Chaudhuri, R., Yang, P., Vafaee, F., Fazakerley, D., Humphrey, S., James, D. & Kuncic, Z., (2016). Unraveling kinase activation dynamics using kinase-substrate relationships from temporal large-scale phosphoproteomics studies. PLoS One, 11(6), e0157763.
2015
Systems biology:31. Pathania, R., Ramachandran, S., Elangovan, S., Padia, R., Yang, P., Cinghu, S., Veeranan-Karmegam, R., Fulzele, S., Pei, L., Chang, C., Choi, J., Shi, H., Manicassamy, S., Prasad, P., Sharma, S., Ganapathy, V., Jothi, R. & Thangaraju, M. (2015). DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nature Communications, 6, 6910. [Pubmed]
30. Hoffman, N., Parker, B., Chaudhuri, R., Fisher-Wellman, K., Kleinert, M., Humphrey, S., Yang, P., Holliday, M., Trefely, S., Fazakerley, D., Stockli, J., Burchfield, J., Jensen, T., Jothi, R., Kiens, B., Wojtaszewski, J., Richter, E. & James, D. (2015). Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metabolism, 22(5), 922-935. [Pubmed]
Methodology and tools:
29. Yang, P.†, Zheng, X., Jayaswal, V., Hu, G., Yang, J. & Jothi, R. (2015). Knowledge-based analysis for detecting key signaling events from time-series phosphoproteomics data. PLoS Computational Biology, 11(8), e1004403. [Full Text], [Github Repo], [CRAN R package]
2014
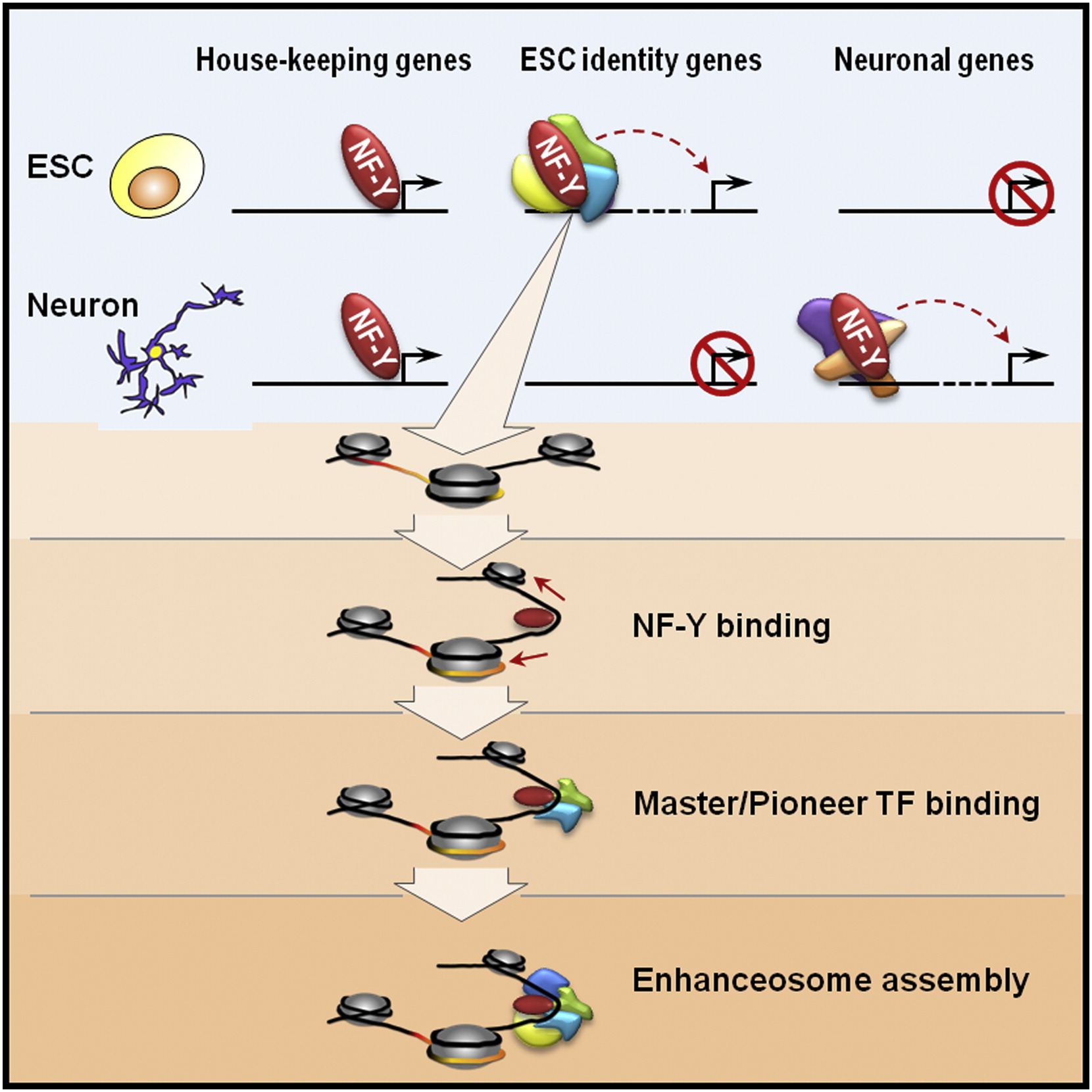
Systems biology:28. Oldfield, A.✢, Yang, P.✢, Conway, A., Cinghu, S., Freudenberg, J., Yellaboina, S. & Jothi, R. (2014). Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Molecular Cell, 55(5), 708-722. [Full Text], [PDF]
27. Ma, X., Yang, P., Kaplan, W., Lee, B., Wu, L., Yang, J., Yasunaga, M., Sato, K., Chisholm, D. & James, D. (2014). ISL1 regulates peroxisome proliferator-activated receptor γ activation and early adipogenesis via bone morphogenetic protein 4-dependent and -independent mechanisms. Molecular and Cellular Biology, 34(19), 3607-3617. [Full text]
26. Lackford, B., Yao, C., Charles, G., Weng, L., Zheng, X., Choi, E., Xie, X., Wan, J., Xing, Y., Freudenberg, J., Yang, P., Jothi, R., Hu, G. & Shi, Y. (2014). Fip1 regulates mRNA alternative polyadenylation to promote stem cell self‐renewal. EMBO Journal, 33(8), 878-889. [Full Text]
Methodology and tools:
25. Yang, P.✢, Patrick, E.✢, Tan, S., Fazakerley, D., Burchfield, J., Gribben, C., Prior, M., James, D. & Yang, J. (2014). Direction pathway analysis of large-scale proteomics data reveals novel features of the insulin action pathway. Bioinformatics, 30(6), 808-814. [Full Text], [PDF], [Github Repo & tutorial], [CRAN R package]
24. Yang, P.†, Yoo, P., Fernando, J., Zhou, B., Zhang, Z. & Zomaya, A. (2014). Sample subset optimization techniques for imbalanced and ensemble learning problems in bioinformatics applications. IEEE Transactions on Cybernetics, 44(3), 445-455. [Full Text] [PDF] [Software]
2013
Systems biology:23. Humphrey, S., Yang, G., Yang, P., Fazakerley, D. J., Stöckli, J., Yang, J. & James, D. (2013). Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metabolism, 17(6), 1009-1020. [Full Text], [PDF]
Methodology and tools:
22. Yang, P., Liu, W., Zhou, B., Chawla, S. & Zomaya, A. (2013). Ensemble-based wrapper methods for feature selection and class imbalance learning. In Proceedings of the 17th Pacific-Asia Conference on Knowledge Discovery and Data Mining (PAKDD), Lecture Notes in Artificial Intelligence 7818, Springer Berlin Heidelberg, 544-555. [Full Text]
21. Yang, P., Yang, J., Zhou, B. & Zomaya. A. (2013). Stability of feature selection algorithms and ensemble feature selection methods in bioinformatics. In Biological Knowledge Discovery Handbook: Preprocessing, Mining and Postprocessing of Biological Data, Wiley, New Jersey, USA, 333-352. [Full Text], [PDF]
2012
20. Yang, P., Humphrey, S., Fazakerley, D., Prior, M., Yang, G., James, D. & Yang, J. (2012). Re-fraction: a machine learning approach for deterministic identification of protein homologues and splice variants in large-scale MS-based proteomics. Journal of Proteome Research, 11(5), 3035-3045. [Full Text], [PDF]19. Yang, P.†, Ma, J., Wang, P., Zhu, Y., Zhou, B. & Yang, J. (2012). Improving X! Tandem on peptide identification from mass spectrometry by self-boosted Percolator. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 9(5), 1273-1280. [Full Text], [PDF]
18. Wang, P., Yang, P. & Yang, J. (2012). OCAP: an open comprehensive analysis pipeline for iTRAQ. Bioinformatics, 28(10), 1404-1405. [Full Text]
2011
17. Yang, P.✢, †, Ho, J.✢, Yang, J. & Zhou, B. (2011). Gene-gene interaction filtering with ensemble of filters. BMC Bioinformatics, 12, S10. [Full Text]16. Yang, P., Zhang, Z., Zhou, B. & Zomaya, A. (2011). Sample subset optimization for classifying imbalanced biological data. In Proceedings of the 15th Pacific-Asia Conference on Knowledge Discovery and Data Mining (PAKDD), Lecture Notes in Artificial Intelligence 6635, Springer Berlin Heidelberg, 333-344. [Text]
2010
15. Yang, P.†, Ho, J., Zomaya, A. & Zhou, B. (2010). A genetic ensemble approach for gene-gene interaction identification. BMC Bioinformatics, 11(1), 524. [Full Text]14. Wang, P., Yang, P., Arthur, J. & Yang, J. (2010). A dynamic wavelet-based algorithm for pre-processing tandem mass spectrometry data. Bioinformatics, 26(18), 2242-2249. [Full Text]
13. Yoo, P., Ho, Y., Ng, J., Charleston, M., Saksena, N., Yang, P. & Zomaya, A. (2010). Hierarchical kernel mixture models for the prediction of AIDS disease progression using HIV structural gp120 profiles. BMC Genomics, 11, S22. [Full Text]
12. Yang, P.†, Zhang, Z., Zhou, B. & Zomaya, A. (2010). A clustering based hybrid system for biomarker selection and sample classification of mass spectrometry data. Neurocomputing, 73(13), 2317-2331. [Full Text]
11. Yang, P.†, Zhou, B., Zhang, Z. & Zomaya, A. (2010). A multi-filter enhanced genetic ensemble system for gene selection and sample classification of microarray data. BMC Bioinformatics, 11, S5. [Full Text]
10. Yang, P., Yang, J., Zhou, B. & Zomaya, A. (2010). A review of ensemble methods in bioinformatics. Current Bioinformatics, 5(4), 296-308. [Full Text], [PDF]
9. Li, L., Yang, P., Ou, L., Zhang, Z. & Cheng, P. (2010). Genetic algorithm-based multi-objective optimisation for QoS-aware web services composition. In Proceedings of the 4th International Conference on Knowledge Science, Engineering and Management (KSEM), Lecture Notes in Computer Science 6291, Springer Berlin Heidelberg, 549-554. [Full Text]
2009
8. Yang, P.†, Xu, L., Zhou, B., Zhang, Z. & Zomaya, A. (2009). A particle swarm based hybrid system for imbalanced medical data sampling. BMC Genomics, 10, S34. [Full Text]7. Zhang, Z., Yang, P., Wu, X. & Zhang, C. (2009). An agent-based hybrid system for microarray data analysis. IEEE Intelligent Systems, 24(5), 53-63. [PDF]
6. Yang, P.† & Zhang, Z. (2009). An embedded two-layer feature selection approach for microarray data analysis. IEEE Intelligent Informatics Bulletin, 10(1), 24-32. [PDF]
5. Yang, P., Tao, L., Xu, L. & Zhang, Z. (2009). Multiagent framework for bio-data mining. In Proceedings of the 4th Rough Sets and Knowledge Technology (RSKT), Lecture Notes in Computer Science 5589, Springer Berlin Heidelberg, 200-207. [Text]
2008
4. Zhang, Z. & Yang, P.† (2008). An ensemble of classifiers with genetic algorithm-based feature selection. IEEE Intelligent Informatics Bulletin, 9(1), 18-24. [PDF]3. Yang, P. & Zhang, Z. (2008). A clustering based hybrid system for mass spectrometry data analysis. In Proceedings of the 3rd Pattern Recognition in Bioinformatics (PRIB), Lecture Notes in Bioinformatics 5265, Springer Berlin Heidelberg, 98-109. [Full Text]
• International student travel award
2. Yang, P. & Zhang, Z. (2008). A hybrid approach to selecting susceptible single nucleotide polymorphisms for complex disease analysis. In Proceedings of BioMedical Engineering and Informatics (BMEI), IEEE, 214-218. [PDF]
