Laboratory of Computational Systems Biology
Deciphering trans-regulatory networks for modelling cell identity and controlling cell-fate decisions
Cells are the fundamental building blocks of all life on earth. In multicellular organisms, these cellular building blocks come in a plethora of types and serve distinct functions to coordinate and support the homeostasis of an organism. Intriguingly, all cell types of an organism share the same genetic code and originate from the same group of pluripotent stem cells that have undergone different cellular fates during development.
Research from the last few decades has established that cell identity and cell-fate decisions are determined by underlying trans-regulatory networks (TRNs) comprised of cell signalling, transcriptional, and epigenetic regulations, and the intra- and extra-cellular networks they form. Understanding how TRNs regulate cell identity and interact through cell-cell communications in a spatial-temporal manner is critical for gaining insight into the complex nature of multicellular development. To this end, our research takes a holistic approach and seeks to reconstruct the TRNs, that cut across multiple molecular programs, for modelling cell identity and controlling cell-fate decisions.
Leveraging on recent advances in single-cell omics technologies and our experitise in machine learning, deep learning, and statistical modelling, we develop computational models to reconstruct TRNs in stem cells and model their differentiation to specialised cell types. By employing a multidisiciplinary approach that combines 'dry' (computation) and 'wet' (laboratory) studies at the systems level, we contribute to the following research aims:
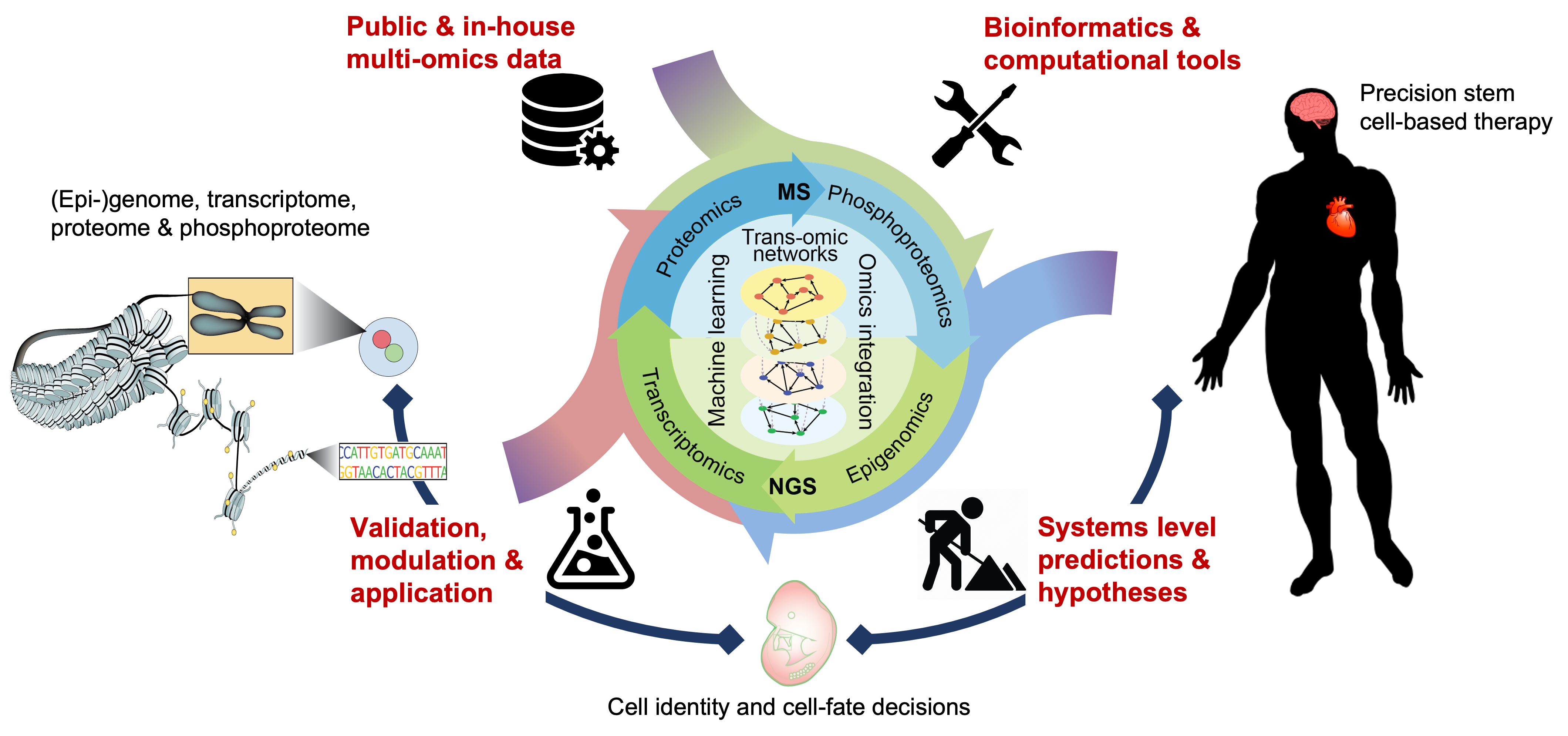
Research from the last few decades has established that cell identity and cell-fate decisions are determined by underlying trans-regulatory networks (TRNs) comprised of cell signalling, transcriptional, and epigenetic regulations, and the intra- and extra-cellular networks they form. Understanding how TRNs regulate cell identity and interact through cell-cell communications in a spatial-temporal manner is critical for gaining insight into the complex nature of multicellular development. To this end, our research takes a holistic approach and seeks to reconstruct the TRNs, that cut across multiple molecular programs, for modelling cell identity and controlling cell-fate decisions.
Leveraging on recent advances in single-cell omics technologies and our experitise in machine learning, deep learning, and statistical modelling, we develop computational models to reconstruct TRNs in stem cells and model their differentiation to specialised cell types. By employing a multidisiciplinary approach that combines 'dry' (computation) and 'wet' (laboratory) studies at the systems level, we contribute to the following research aims:
- Develop computational methods to reconstruct TRNs for modelling cell identity.
- Design computational and experimental protocols to modulate TRNs for guiding cell fate-decisions.
- Gain knowledge on how do different layers of TRNs coordinately regulate cell identity and cell fate-decisions.
- Develop computational framework to assess fidelity of cell conversion for applications such as tissue engineering and stem cell therapy.
