Research summary
Cells are the fundamental building blocks of all life on earth. In multicellular organisms, these cellular building blocks come in a plethora of types and serve distinct functions to coordinate and support the homeostasis of an organism. Intriguingly, all cell types of an organism share the same genetic code and originate from the same group of pluripotent stem cells that have undergone different cellular fates during development.
Research from the last few decades has established that cell identity and cell-fate decisions are determined by underlying molecular programs such as gene transcription and translation, and the intra- and extra-cellular networks they form. Understanding how such programs/networks regulate and interact with one another in a spatial-temporal manner is critical for gaining insight into the complex nature of multicellular development. To this end, our research takes a holistic approach and seeks to understand the molecular 'trans-regulatory networks', comprised of cell signalling, transcriptional, translational, and (epi)genomic regulations, in controlling cell identity and cell-fate decisions. In particular, we develop computational and statistical models to reconstruct each layer of the trans-regulatory networks (e.g. signalling networks, transcriptional networks) and characterise their cross-talk in stem cells during their differentiation to specialised cell types. By employing a multidisiciplinary approach that combines 'dry' (computation) and 'wet' (laboratory) studies at the systems level, we aim to address the following research questions:
- How do different layers of trans-regulatory networks coordinately regulate cell identity and cell fate-decisions?
- How can we accurately predict cell identity and cell fate-decisions based on their trans-regulatory networks?
- How can we modulate trans-regulatory networks to direct cell fate-decisions for applications such as stem cell therapy?
Mapping and modelling trans-regulatory networks in stem cells and progenitor cells for understanding development, tissue regeneration, and stem cell therapy
The ability of stem and progenitor cells to self-renew and differentiate into specialised cell types in the body makes them the key cellular tools for studying normal development, developmental diseases, and for creating stem cell-based therapy, whereby specific cell populations, tissues, and organs compromised by disease, injury or ageing can be replaced and replenished indefinitely.A major initiative in our group is to integrate multi-omics datasets generated by state‐of‐the‐art mass spectrometer (MS) and next generation sequencer (NGS) from various stem and progenitor cells and during their differentiation to specialised cell types. The goal is to reconstruct trans-regulatory networks and to understand how different regulatory machineries (e.g. signalling, transcription, and epigenomics) cooperate to define cell types, functions, and fates, and to translate these knowledge for directed cell type differentiation and tissue/organ regeneration.
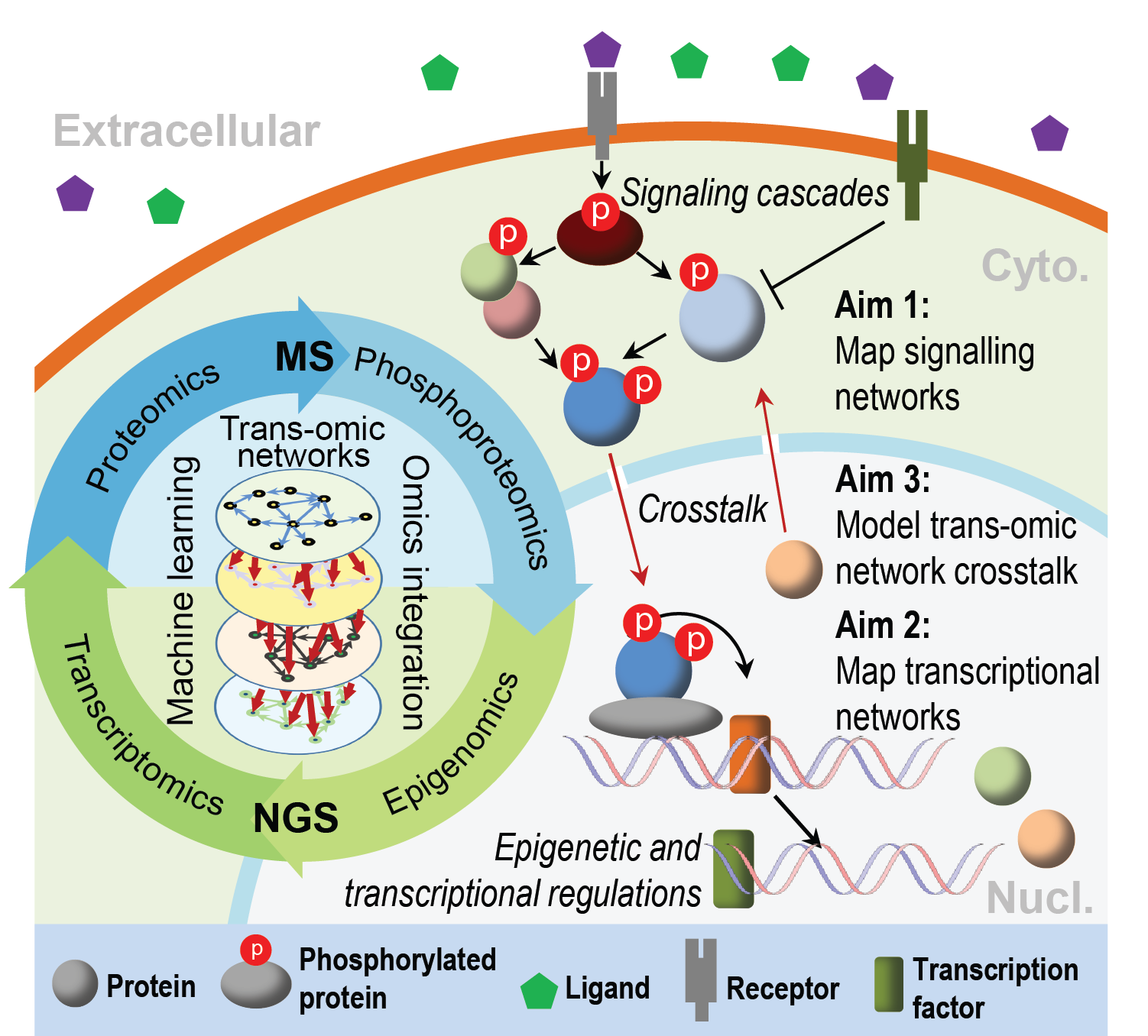
Reconstructing trans-regulatory networks in mouse embryonic stem cells (ESCs)
Pluripotency defines the potential of pluripotent stem cells (PSCs) to differentiate into all somatic and germ cells. We have previously profiled the multi-layered trans‐omics networks in ESCs during their pluripotency transition (Yang et al. 2019). Building on this work, we are developing methods to characterise signaling cascades, transcriptional networks, and translational protein networks in regulating the transition of pluripotency in ESCs, and to uncover how information is passed on from each of these regulatory layer to the next layer. This project will provide a unique opportunity for developing computational and experimental methods for comprehensive understanding of stem cell systems and their decision-making process.
'Mini organs' in a dish: identifying molecular controls in generating brain and retina organoids from human PSCs
While tremendous progress has been made in stem cell therapy, attempts to direct stem/progenitor cells to clinically useful cell types have often been fraught with imprecise and incomplete differentiation. In collaboration with Dr. Anai Gonzalez Cordero's group at CMRI, we are reconstructing trans-networks networks and identifying key network modules and nodes that underlie the generation of brain and retina organoids using human PSCs. Discovery made from these works will have provide both basic and translational knowledge for directed cell type differentiation and tissue/organ regeneration.
Identifying molecular controls in muscle cell formation
In collaboration with Dr. Benjamin Parker's group at the Melbourne University, we are identifying molecular controls that regulate the differentiation of progenitor cells to mature muscle cells. To do this, we have profiled the trans-regulatory networks on phosphoproteomics and proteomics levels and have developed various computational methods allowing the identification of key phosphorylation events and their downstream transcriptional regulation in controlling myogenesis, formation of muscle cells (Xiao et al. 2019). Further exploration on the transcriptomics and epigenomics levels is needed to link the signalling regulation with downstream processes. Finding from this study will have implications in muscle tissue regeneration.
Interrogating cell identity and cell-fate decisions in spatial contexts and using single-cell multi-omics
The advance of single-cell omics and its recent development towards multimodality, that allows profiling of gene expression, chromatin state, protein abundance and spatial location, creates unprecedented opportunities to study complex biological systems at resolutions that were previously unattainable. Our group is harnessing the power of these new biotechnologies to study how each single cell acquires its identity in organs and tissues and makes decisions during cell differentiation and embryogenesis.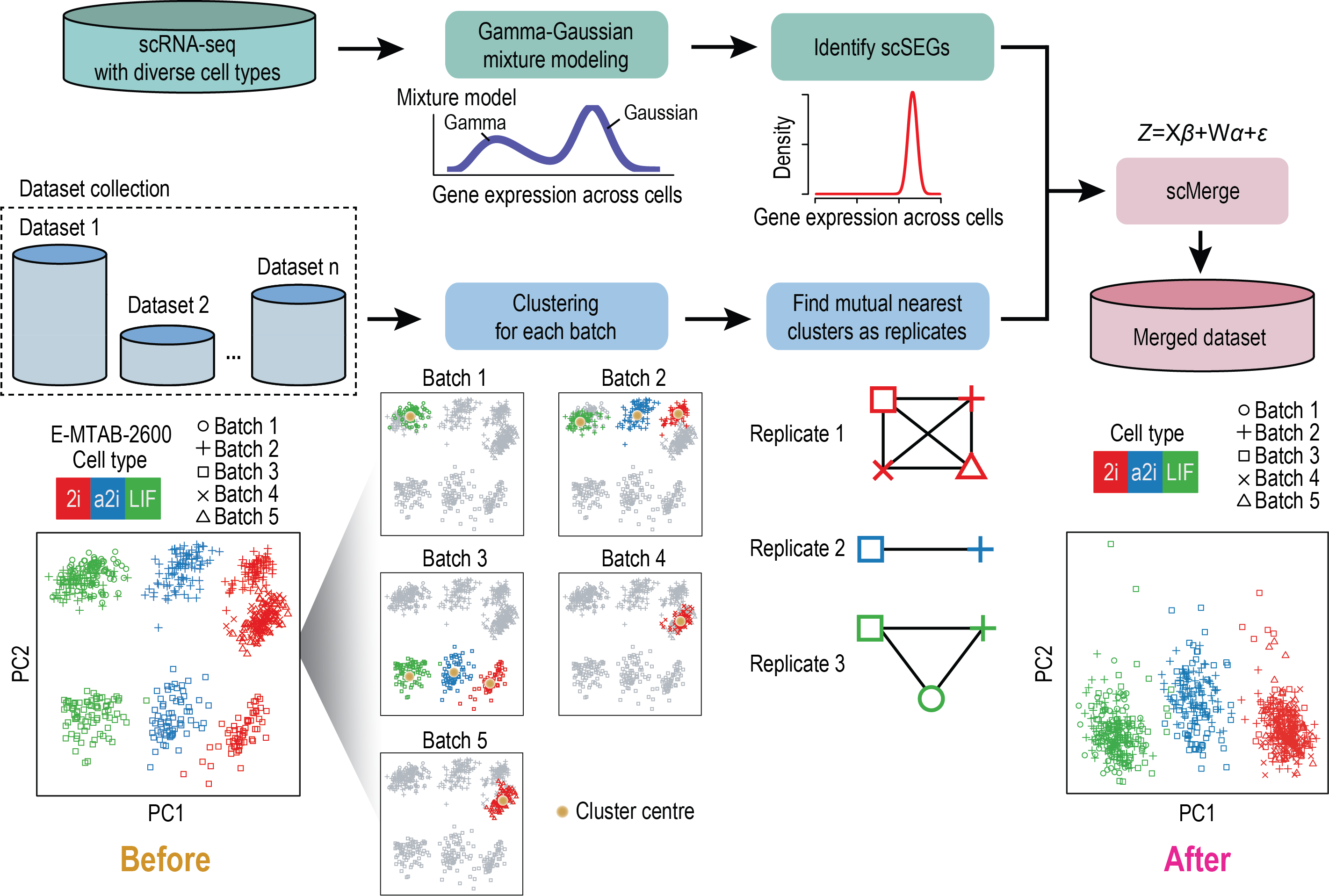
Creating a retina cell type altas using single-cell data acquired from primary retina tissues and retina organoids
We are collaborating with Dr. Anai Gonzalez Cordero's group at CMRI to create the first retina cell type atlas by integrating single-cell omics data generated from various primary retina tissues and retina organoids. From this atlas, we uses our recently developed Cepo algorithm to derive a cell identity score for each gene with respect to the cell types in the atlas (Kim et al. 2021). These cell identity scores provide a basis for annotating cell types and assessing data quality and disease status in future single-cell omics data generated from retina tissues and retina orgnoids.
Identifying cells that show bipotential differentiation capacity during early embryogenesis
This project involves experimental profiling of single cells in embryos during their differentiation to multiple cell lineages. Together with Prof. Patrick Tam's lab, we are developing experimental and computational techniques for uncovering cells that could differentiate into multiple lineages during early embryogenesis. The results from this study will provide new knowledge to cell lineage specification in embryogensis and shed light on early developmental diseases
Developing computational methods for pre-processing, analysing, and integrating bulk and single-cell multi-omics data and their spatial information
Much of the work in the above research direction requires computational methods that are capable of extracting biological knowledge from large-scale of omic data generated from bulk and single-cell omics technologies. To enable these biological interrogation and discoveries, we specialise in developing computational and statistical machine learning models for analysing and integrating these data.Machine learning methods for cell type annotation using single-cell multi-omic data
Cell type identification is a key step in single-cell omics experiments. Our previous work has demonstrated that machine learning could be effectively applied to annotate cell types using their transcriptomic profiles (Lin et al.). Building on this work, we are in process of developing deep learning-based methods for annotating cell types and identifying molecular features associated with cell type annotation from single-cell multi-omics data.
Estimating the number of cell types from single-cell omic data
Identifying the number of cell types in a biological system is an essential task prior in cell type annotation. Building on our previous work that uses an ensemble of deep learning models for cell type clustering (Geddes et al.), we have recently implemented a stability based approach for number of cell type estimation (Yu et al.) from single-cell transcriptomics data. We are now exploring their application in single-cell multi-omics data.
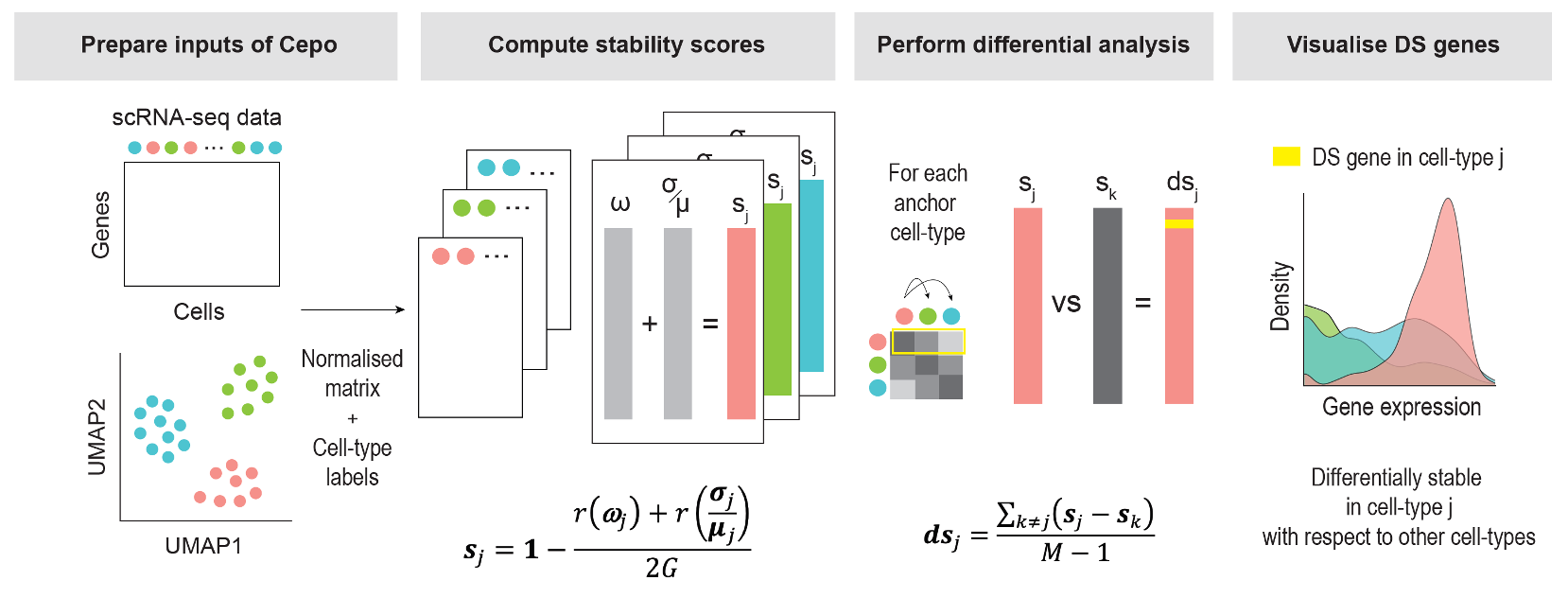
Developing computational methods for detecting cell identity genes
We have recently proposed a computational framework that uses differential stability of gene expression between cell types to detect for cell identity genes (Kim et al. 2021). We are working on extending this approach for detecting genes that mark the changes in cell status such as in healthy individual and diseases. This work will allow us to pinpoint the genes that mark disease status in a cell type and may provide knowledge for potential treatment.
Openings
Post-doctoral positions funded by NHMRC grant are available for conducting research on the broad area of computational systems biology in stem cells and their therapeutic application. Interested candidates are encouraged to please contact Pengyi Yang, lab head, to discuss potential projects and other details (pengyi.yang [at] sydney.edu.au)PhD scholarships are available for both domestic and international candidates. Please see link for more details on Children's Medical Research Institute PhD Research Award. For details on scholarships offered at the University of Sydney, please see link.
Honours projects are available through either Children's Medical Research Institute, Faculty of Health and Medicine, see link; or the School of Mathematics and Statistics, Faculty of Science. For more details regarding projects, please contact Pengyi Yang.
Summer research scholarships are available for third year undergraduates at both Children's Medical Research Institute and Charles Perkins Centre. For more details regarding potential projects and scholarships, please contact Pengyi Yang.
For postgraduate applicants, all 'wet' projects require experience with cell culturing and good understanding of basic experimental protocols (e.g. qPCR, westernblotting) and preferably experience of (single cell-)sequencing and mass spectrometry-based proteomics. All 'dry' projects require programming skill in at least one programming/scripting language (R, Perl, Python, Java, C++, C and Matlab). There are ample opportunity to work closely with biologists, statisticians, bioinformaticians, and computer scientists.
More information about scholarship opportunity at the University of Sydney can be found here.
For obtaining scholarships and top-ups from CMRI, please read here.